Genetics and Genomics of Leiomyosarcoma (LMS): Improved understanding of cancer biology and new approaches to diagnosis and treatment
University of Michigan
Principal Investigator(s):

Scott M. Schuetze, MD, PhD

Jonathan A. Fletcher, MD
- Principal Investigator(s) Contact Information
- Overview
- Project 1: Genomic Vulnerabilities in Leiomyosarcoma (LMS)
- Project 2: Understanding and exploiting the genetic basis of LMS development
- Project 3: Applying Liquid Biopsy Technologies to Detect Clinical Response and Mechanisms of Resistance in the Treatment of LMS
- Administrative Core
- Biospecimen Core
- Data Analysis Core
- Developmental Research Program
- Career Enhancement Program
- Institutional SPORE Website
Principal Investigator(s) Contact Information
Scott M. Schuetze, MD, PhD
University of Michigan
1500 E Medical Center Dr.
Rogel Cancer Center
Floor B1 Reception E
Ann Arbor, MI 48109
Tel: 734-647-8921
Jonathan A. Fletcher, MD
Associate Professor of Pathology
Brigham and Women’s Hospital, Harvard Medical School
20 Shattuck Street, Thorn 528
Brigham and Women’s Hospital
Boston, MA 02115
Tel: 617-732-5152
Mobile: 617-816-4142
Overview
This SPORE grant is entitled Genetics and Genomics of Leiomyosarcoma (LMS): Improved understanding of cancer biology and new approaches to diagnosis and treatment. This SPORE brings together a stellar team of experts who have been effectively working together over the past 4 years despite spanning institutions across the US, Canada, and Australia.
Leiomyosarcoma is a rare cancer. The estimated annual occurrence rate is 1700 new patients. We recognized that to effectively study a rare cancer we had to include several outstanding cancer centers: University of Toronto, The Ohio State University, Mayo Clinic, Huntsman Cancer Institute, DFCI/BWH at Harvard, MSKCC, Garvan Institute/University of New South Wales, Cold Spring Harbor National Laboratory and the University of Michigan. Our goal is to improve the knowledge regarding leiomyosarcoma (LMS) genetics, biology, and therapeutic approaches to rationally develop novel and more effective therapies including combinations of different agents targeting different pathways to exploit unique vulnerabilities. Our program includes 3 projects, 3 cores (Administrative, Biospecimen and Data Analysis) and 2 programs (Career Enhancement and Developmental Research).
Project 1 aims to identify and exploit genomic vulnerabilities in LMS targeting the dependency on the non-homologous end joining DNA repair pathway and the associated enzyme DNA-activated protein kinase (DNA-PK). Project 2 focuses on studying the genetic epidemiology of LMS defining the risk for cancer in families with cancer predisposition syndromes such as Li-Fraumeni Syndrome, as well as the general population by evaluating the largest ever cohort of LMS patients. The objectives of Project 3 are to collect and analyze serial liquid biopsy samples from blood samples of more than 200 patients with metastatic LMS enrolled in a randomized prospective clinical trial for circulating tumor DNA (ctDNA) to monitor clinical outcomes and to characterize and validate recurrent patterns of tumor heterogeneity and tumor evolution including the emergence of resistance.
The Developmental Research Program is designed to identify and support novel projects focused on translational research related to leiomyosarcoma. The Career Enhancement Program has clearly delineated processes for solicitation and selection of awardees from diverse backgrounds, with strong mentoring that guarantee the successful transition into translational research careers.
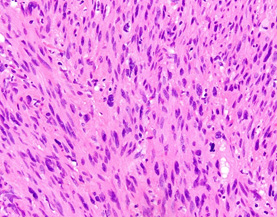
Image courtesy of Jason Hornick, MD, PhD, Professor of Pathology, Harvard Medical School
Project 1: Genomic Vulnerabilities in Leiomyosarcoma (LMS)
Project Co-Leaders:
Jonathan Fletcher, MD (Basic)
Adrian Marino-Enriquez, MD, PhD (Basic)
Suzanne George, MD (Clinical)
LMS have pronounced, heterogeneous and unstable chromosomal aberrations that provides opportunities for studies of drugs targeting chromosomal instability. LMS have homologous recombination (HR)-deficiency with associated hyper-dependency on the compensatory non-homologous end joining (NHEJ) DNA repair pathway. We have shown that LMS are highly dependent on the NHEJ protein, DNA-activated protein kinase (DNA-PK). TP53 inactivation results in tolerance to aneuploidy and double-strand DNA damage, and concurrent RB1 inactivation exacerbates these defects, hastening acquisition of tumor suppressor gene mutations. We have shown TP53 and RB1 inactivation in 90% of LMS, and TP53 and RB1 inactivating germline mutations are initiating events in LMS.
Project 1 seeks to develop therapies that maximize intrinsic LMS genotoxic stress by targeting the key DNA damage response enzyme, DNA-PK. Our preliminary studies show that DNA-PK inhibition increases dsDNA breaks in LMS. This genotoxic damage was compounded by pre-treating or co-treating the cells with a DNA-damaging agent (doxorubicin). A genome-wide shRNA long-term screen identified genes with essential roles in early-passage LMS lines. DNA-PK (gene name PRKDC) is among the top 0.5% of essential genes in LMS, and in dedifferentiated liposarcoma (another common sarcoma subtype with CIN). Likewise, the essential DNA-PK substrate, RPA2, ranked in the top 2% of essential genes in LMS. These observations are the rationale for the overarching Project 1 hypothesis, which is that the intrinsic CIN in advanced LMS is a vulnerability that can be exploited, using doxorubicin coupled with targeted inhibitors, particularly DNA-PK inhibitors.
It is important to highlight the rigor of the laboratory investigation including the mechanistic work using our well-annotated and published cell lines, CRISPR experiments, and preclinical validation of therapies leading to clinical trials targeting LMS hyperdependencies.
Specific Aim 1: To characterize CIN, DDR, DNA-PK activation, and TP53 status heterogeneity within and between LMS metastases in a given patient.
Specific Aim 2: To characterize DNA-PKi response/resistance mechanisms and synthetic lethalities in LMS.
Specific Aim 3: To evaluate genotoxic therapies for LMS in preclinical models and clinical trials.
A Phase 1/2 clinical trial of low-dose doxorubicin combined with peposertib (DNA-PKi) will formally test our hypothesis and evaluate this approach as effective LMS treatment.
Project 2: Understanding and exploiting the genetic basis of LMS development
Project Co-Leaders:
David Thomas, MD, PhD (Basic)
Judy Garber, MD, MPH (Clinical)
Joshua Schiffman, MD (Clinical)
We are conducting the first comprehensive genetic epidemiology study to focus on leiomyosarcoma. TP53 is somatically mutated in almost all LMS. Germline mutation found in 10-15% of patients with LMS is a feature of Li-Fraumeni Syndrome (LFS). The project will identify germline pathogenic variants (PVs) in TP53, and other DNA damage repair (DDR) genes in patients with LMS. Project 2 also seeks to identify previously unrecognized genes and pathways conferring risk of LMS development.
We will evaluate a large cohort of LMS patients to identify the roles of cancer predisposition genes using WGS to define the genetic factors associated with LMS risk. This definitive genetic map will facilitate counselling, screening of at-risk relatives, risk management and identify new cancer targets. We are also collaborating with the largest cohort to date of patients with LFS to uniquely explore genotype/phenotype associations. This analysis will allow us to define the risk for LMS in this population. Our hypothesis is that comprehensive assessment of germline DNA of cancer predisposition genes in patients with LMS and LFS will identify families at increased risk of LMS who may benefit from enhanced cancer screening.
Specific Aim 1: To estimate the risk for cancers in families of patients with LMS. Our team will interrogate two complementary assets: the Utah Population Database (UPDB), and the International Sarcoma Kindred Study (ISKS) to estimate the risk-to-relatives of LMS patients of LFS- or cancer predisposition syndrome-related cancers.
Specific Aim 2: To examine germline pathogenic variants in known cancer predisposition genes and pathways, to identify new susceptibility genes, and to evaluate attitudes of genetic testing in LMS.
Specific Aim 3: To characterize LMS risk in patients with known TP53 germline PVs. We include data from the LiFE Consortium, a germline TP53 specific registry used by the LFS community
The results from Project 2 will reveal important insights into the genetic basis of LMS, directly impact future risk management and treatment of patients at risk for LMS, and establish a global infrastructure to enable future research into the genetic basis for this disease.
Project 3: Applying Liquid Biopsy Technologies to Detect Clinical Response and Mechanisms of Resistance in the Treatment of LMS
Project Co-Leaders:
Brian Crompton, MD (Basic)
Scott Schuetze, MD, PhD (Clinical)
Brittany Siontis, MD (Clinical)
The treatment of advanced leiomyosarcoma (LMS) includes the use of chemotherapy. Only 15-20% of patients experience LMS response to chemotherapy, but nearly all experience toxicity. Biomarkers predicting response do not yet exist for LMS, preventing clinicians from recommending treatment only for patients most likely to benefit. Sensitive measures of treatment response would allow physicians to limit therapy and associated toxicities in patients who are unlikely to benefit from a chemotherapy.
Liquid biopsies are new technologies that allow detection of ultra-rare tumor fragments in body fluids. Circulating tumor DNA (ctDNA) can be identified in plasma drawn at the time of diagnosis. Changes in ctDNA levels in the plasma correlate with changes in disease burden. We’ve confirmed that patients with LMS have detectable levels of ctDNA at the time of diagnosis and treatment. Applying this approach to patients with LMS could result in the validation of ctDNA as a biomarker of prognosis and as an early measure of response to chemotherapy.
Hypothesis: ctDNA may be used as a biomarker of LMS patient prognosis and tumor response to chemotherapy. We will collect and analyze serial liquid biopsy and tumor samples to validate ctDNA plasma levels as a biomarker in patients with metastatic LMS and to perform deep genomic profiling of this tumor DNA to describe recurrent patterns of tumor evolution and treatment resistance in this disease.
Specific Aim 1: To determine the association of ctDNA levels and recurrent copy-number alterations with outcome in patients with metastatic LMS treated with first-line chemotherapy.
Specific Aim 2: To characterize and validate recurrent patterns of tumor heterogeneity and tumor evolution in patients with metastatic LMS undergoing chemotherapy.
At the conclusion of this project, we anticipate that we will have validated the first dynamic biomarker of prognosis and treatment response for clinical use in patients with LMS. Such a tool is expected to allow patients to optimize their treatment choices by justifying potential exposures to side effects only for patients who are likely to respond and by identifying patients who are very unlikely to experience benefit from chemotherapy who may choose to pursue other experiment treatment.
Administrative Core
Core Directors:
Scott Schuetze, MD, PhD
Denise Reinke, MS, NP, MBA
The Administrative Core facilitates this SPORE’s research in leiomyosarcoma (LMS) expediting the translation of discoveries into new concepts of cancer biology and genetics as well as methods of detection and treatment. This core coordinates the many activities required to maximize this research effort. Our three Project Teams (Basic and Clinical) are located at Dana Farber Cancer Institute (DFCI), Garvan Institute of Medical Research (Australia), Huntsman Cancer Institute, Mayo Clinic, and the University of Michigan. The leadership of our cores and programs are from the University of Michigan, BWH/DFCI, Research Institute at Nationwide Children’s Hospital, the University of Toronto and Weill-Cornell. An Executive Committee has been formed to engage project, core, and program leadership. The Executive Committee is chaired by Judy Garber, MD, MPH, Director of the Center for Cancer Genetics and Prevention at Dana-Farber Cancer Institute. The Principal Investigators have convened a strong External Advisory Board (EAB). The EAB is comprised of experts in several cancer research fields including cancer center directors and SPORE PIs. The EAB will meet annually to review progress as well as review and critique plans for the upcoming year. David Tuveson, MD, PhD, Director of the Cold Spring Harbor Laboratory Cancer Center, chairs our EAB.
Biospeciman Core
Core Directors:
Jason Hornick, MD, PhD
Nilsa C. Ramirez, MD
The Biospecimen Core will provide expert pathology review including expert IHC analysis to confirm diagnosis and provide centralized, specialized services, including nucleic acid extraction, digital slide imaging, standardized collection, receipt, processing, storage, and distribution of biospecimens prospectively collected on the LMS SPORE trials and studies. The Core begins with histologic and IHC confirmation of the diagnosis of leiomyosarcoma. The specific challenge in LMS diagnosis is amplified with the phrase that the predominant cell “resembles” a smooth muscle cell. Indeed, accurate IHC staining is required to support the diagnosis.
The Biospecimen Core is housed at the Surgical Pathology Lab and the IHC lab at Brigham and Women’s Hospital for pathology confirmation and at the Biopathology Center (BPC), at Nationwide Children’s Hospital in Columbus, Ohio, which will provide a full range of services related to biospecimen procurement, banking, processing, and distribution. Both facilities are fully accredited.
Figure 1: LMS SPORE Biospecimen Core
Data Analysis Core
Core Directors:
Veera Baladandayuthapan, PhD
Benjamin Haibe-Kains, PhD
Karla V. Ballman, PhD
The Data Analysis Core (DAC) will advance the understanding of leiomyosarcoma by providing a centralized and comprehensive resource for study design, biostatistics, bioinformatics, data collection and management, data analytics, and data dissemination. The DAC supports all SPORE research activities including the individual projects, the Developmental Research Program [DRP] and Career Enhancement Program [CEP], the other SPORE Cores, and pan-SPORE endeavors. These projects require a breadth of analytic and computational methodologies. The biostatistical and bioinformatics of each project vary significantly, and the expertise of the DAC members allows the optimal design for the experimental/clinical studies and generation of an appropriate analysis plan determined collaboratively with the project investigators. The DAC will advance the goals of the three research projects, the DRP, CEP, and the Biospecimens Core through the following: (1) thoughtful biostatistical and bioinformatics collaboration that yields efficient study designs, thorough and appropriate analyses, quality data management, and timely dissemination of results, (2) implementation of best practices for data collection, management, integration, and storage, and (3) development of resourceful methodology that enhances the research across the SPORE projects.
Developmental Research Program
Program Director:
Steven Robinson, MBBS
The mission of the DRP is to foster new endeavors targeting leiomyosarcoma (LMS) or soft tissue sarcomas of high genomic complexity; promote essential technologies currently unavailable within our Cores; and advance promising concepts from pilots to sustainable projects. To these ends, the DRP will provide short-term funding for meritorious projects that break new ground in sarcoma research. The DRP will particularly prioritize projects most likely to thrive within the translational and multidisciplinary setting of the SPORE.
Career Enhancement Program
Program Directors:
Elizabeth G. Demicco MD, PhD
Jonathan A. Fletcher MD
The overall goal of the SPORE Career Enhancement Program (CEP) is to bring talented researchers to the area of sarcoma translational research. Given the focus of this SPORE is leiomyosarcoma (LMS), the CEP will seek applicants whose research addresses translational challenges in LMS or other sarcomas with complex genomic and biologic features, including undifferentiated pleomorphic sarcoma, myxofibrosarcoma, or dedifferentiated liposarcoma as well as research focused on translational DNA damage repair research, or molecular diagnostics. This program will provide the opportunity for investigators to develop their potential as independent translational researchers. It is a priority of the SPORE CEP to expand the personal and professional diversity of highly qualified investigators in the field of sarcoma, to bring new perspectives and research interests to the field.







