SPORE in Soft Tissue Sarcoma
Memorial Sloan Kettering Cancer Center
Principal Investigator(s):

Samuel Singer, MD
- Principal Investigator(s) Contact Information
- Overview
- Project 1: Molecular Determinants of GIST Pathogenesis
- Project 2: Targeting Hippo Pathway Dependence in Genetically Complex Sarcomas
- Project 3: Epigenetic and Immuno-Oncologic Vulnerabilities in Synovial Sarcoma
- Administrative Core
- Biospecimen Repository Core
- Biostatistics and Bioinformatics Core
- Clinical Trials and Advocacy Core
- Developmental Research Program
- Career Enhancement Program
Principal Investigator(s) Contact Information
Samuel Singer, MD
Chief, Gastric and Mixed Tumor Service
Director, MSK Sarcoma Center
Vincent Astor Chair of Clinical Research
Memorial Sloan Kettering Cancer Center
1275 York Avenue
New York, NY 10065
(212) 639-2940
Overview
The SPORE in Soft Tissue Sarcoma seeks to reduce morbidity and mortality from soft tissue sarcoma by developing therapies targeted to specific molecular, genetic, epigenetic, and signaling pathway alterations or specific sarcoma type and subtype. The SPORE’s 3 translational research projects each aim to identify new therapeutic targets via defining molecular mechanisms of sarcomagenesis and of resistance to targeted therapies, to clinically validate those targets in patient samples, establish new clinical trials, and discover predictors of outcome and response to targeted therapy. Our team leverages a unique resource, a clinicopathologic and outcomes database including over 15,000 patients treated for soft tissue sarcoma at MSK, linked to an extensive sarcoma tissue/blood bank, genetic and epigenetic data, and primary sarcoma cell lines and xenograft models.
Project 1: Molecular Determinants of GIST Pathogenesis
Project Co-Leaders:
Ping Chi, MD, PhD (Basic Co-Leader)
Cristina Antonescu, MD (Clinical Co-Leader)
Gastrointestinal stromal tumor (GIST), which arises in nerve cells located in the walls of the digestive system, is one of the most prevalent sarcoma subtypes. Advanced unresectable and/or metastatic GISTs are rarely curable despite initially being controlled by current treatments. Current tools for predicting prognosis underestimate the likelihood of the cancer coming back for some cases, and no current prediction tools consider molecular biomarkers. We have found that mutations that inactivate the genes MAX or MGA (which occur in 5% of GISTs) on both chromosomes and extra copies of the pro-cancer gene MYC (0.5%) are more common in untreated metastatic GIST compared to primary localized GIST, and, along with deletion of one arm of chromosome 1, are associated with greater risk of relapse. We hypothesize that these genetic changes promote tumor formation and aggressive metastatic behavior by preventing a key epigenetic regulator from working and may represent independent molecular risk biomarkers for GIST relapse. Here, we propose studies of cancer models and patient tumors using cutting-edge analysis of the arrangement and readability of genes and their activity, combined with computational methods, to 1) elucidate how MAX/MGA/MYC mutations affect GIST development, and 2) identify and validate molecular biomarkers predictive of recurrence risk. These studies could change how treatments are selected for GIST patients and identify new ways to treat GIST metastases.
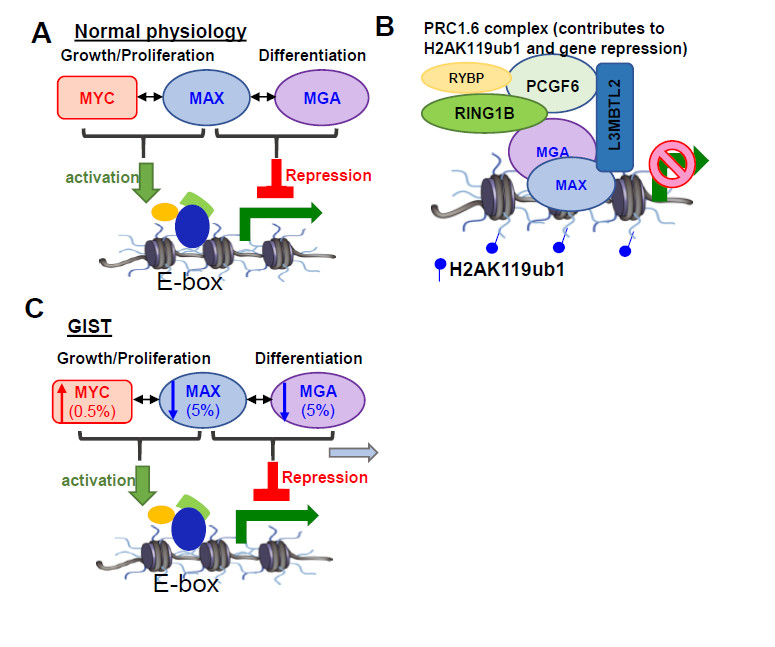
MAX/MGA/MYC mutations affect GIST development
Project 2: Targeting Hippo Pathway Dependence in Genetically Complex Sarcomas
Project Co-Leaders:
Samuel Singer, MD (Clinical Co-Leader)
Hans Guido Wendel, MD (Basic Co-Leader)
Our overall goal is to find effective therapies for 4 of the most common and clinically aggressive types of sarcomas that arise because of diverse combinations of mutations and extra or missing copies of genes: well-differentiated liposarcoma (WDLS), dedifferentiated liposarcoma (DDLS), myxofibrosarcoma (MFS), and undifferentiated pleomorphic sarcoma (UPS). These sarcomas often come back or spread to other organs after surgery, and patients have few treatment options once they develop advanced disease. While the complexity of genetic changes in these sarcomas has made it difficult to determine which are most responsible for helping these tumors develop and spread, we have found that all 4 subtypes share genetic changes that cause increased signaling through a group of interacting proteins called the Hippo pathway, which control cell proliferation, cell death, and stem cell renewal. These genes’ translation into protein requires a specific enzyme to unwind their RNA called eIF4A. We therefore hypothesize that most WDLS, DDLS, MFS, and UPS tumors depend on Hippo signaling and eIF4A for growth and survival. In this project, we will first examine the role of the Hippo pathway in promoting sarcoma formation, controlling specification of cells into specific tissues, tumor progression, and metastasis. Second, we will evaluate the efficacy of the new eIF4A inhibitor TDI-7663 both in cell lines and in 182 mouse models bearing genetically diverse patient tumors. The mouse experiments will also allow a rigorous study of how tumors become resistant to eIF4A inhibition. We will map the effects of TDI-7663 on RNA translation in sarcoma cells and mouse models. Finally, we will advance TDI-7663 toward the clinic by determining its toxicity, pharmacology, and efficacy, which will inform an application for investigative new drug (IND) status with the FDA. Clarification of the roles of the Hippo pathway and cancer-promoting RNA translation will help us understand how these sarcomas form and spread, identify new drug targets, identify effective combination therapies to combat resistance, and enable precision oncology. We expect that the IND-enabling studies will lead to a phase 1 clinical trial of TDI-7663 in patients with DDLS, MFS, and UPS within 5 years.
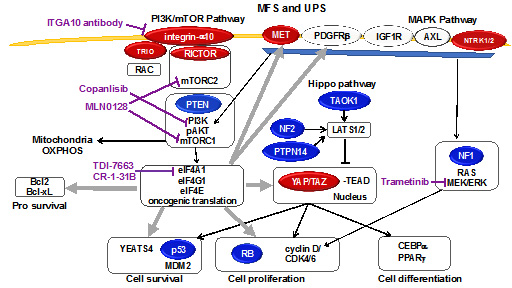
Model for tumorigenic Hippo signaling in MFS/UPS. Red, oncoproteins amplified in sarcomas. Blue, tumor suppressors deleted in sarcomas. Wide gray arrows, translational regulation. Purple, inhibitors.
Project 3: Epigenetic and Immuno-Oncologic Vulnerabilities in Synovial Sarcoma
Project Co-Leaders:
Marc Ladanyi, MD (Clinical Co-Leader)
Christopher Klebanoff, MD, PhD (Basic Co-Leader)
Sandra D’Angelo, MD (Clinical Co-Leader)
Synovial sarcoma (SS) is caused by dysregulation of how genes are arranged and chemically modified to control their accessibility to be read as a result of fusion of genes encoding two proteins that help control these processes: SS18 and SSX. Understanding how gene architecture and accessibility, or epigenetics, is changed in SS might suggest specific ways that drugs could be used to restore normal genetic control, which could work well in combination with immunotherapy. At the same time, gene fusions such as SS18::SSX may generate unique protein fragments that trigger immune responses against tumors, called neoantigens, because they differ significantly from normal, non-tumor proteins. Thus, our overall goal is to identify epigenetic and immuno-oncologic vulnerabilities in SS and potential synergies between them toward treatments that control metastatic SS. The first goal of this project is to decipher how loss of a specific epigenetic regulator, SETD2, changes (a) patterns of a specific chemical modification across all genes and (b) the reading of genes in SS cells and patient tumors. We will also determine whether drugs that act on epigenetic regulators restore normal patterns and explore whether loss of SETD2 affects how well the immune system can attack SS. The second goal is to measure the degree to which the SS18::SSX fusion is recognized by a specific class of immune cells (T cells) and to test whether T cells genetically engineered to recognize the SS18::SSX fusion can shrink or control SS tumors. The third goal is to determine the tolerability, immune effects, and efficacy of a vaccine consisting of RNA encoding the SS18::SSX junction and other proteins recognized by T cells that are highly expressed in SS, in combination with an immunotherapy that blocks inhibition of T cells, in a phase I/II clinical trial.
Administrative Core
Core Director:
Samuel Singer, MD, Director
The Administrative Core manages and coordinates the activities of the SPORE program by providing administrative direction and structure, fiscal and programmatic oversight, and coordination with internal and external advisory committees. The Core also assists the directors of the Developmental Research and Career Enhancement Programs in administering those programs.
Biospecimen Repository
Core Directors:
Cristina R. Antonescu, MD, Co-Director
Narasimhan P. Agaram, MBBS, Co-Director
The Biospecimen Repository Core coordinates the collection, annotation, storage, distribution, and tracking of tissue and blood biospecimens from soft tissue sarcoma patients enrolled in research protocols, and ensures linkage of those samples with clinical data in collaboration with project leaders, the bioinformatics core, and the administrative core. The Core also provides SPORE investigators with expert histopathological evaluation of tumor samples from both patients and genetically engineered and xenograft models and assists in performing and interpreting immunohistochemical and in situ hybridization assays.
Biostatistics and Bioinformatics
Core Directors:
Nicholas Socci, PhD, Co-Director
Li-Xuan Qin, PhD, Co-Director
The Biostatistics and Bioinformatics Core supports investigators of the Soft Tissue Sarcoma SPORE in the computational and statistical aspects of their research efforts, including the design, analysis, and interpretation of laboratory experiments, molecular profiling assays, and clinical trials. For analyses for which current methodologies are inadequate, Core staff develop new, alternative methods. The Core also facilitates the management and updating of the clinical sarcoma database in collaboration with the leaders of research projects and the other 3 cores.
Clinical Trials and Advocacy
Core Directors:
William D. Tap, MD, Co-Director
Julia Glade Bender, MD, Co-Director
The Clinical Trials and Advocacy Core facilitates the establishment and conduct of novel clinical trials in soft tissue sarcoma that span age groups. CF3 bridges the adult and pediatric sarcoma programs at MSK; the basic science, translational, and clinical aspects of the various SPORE projects; the Administrative, Biospecimen, and Biostatistics and Bioinformatics Cores; and other Sarcoma SPOREs. Critically, CF3 assists SPORE investigators in acquiring funding for clinical trials through cooperative/government funding mechanisms, pharmaceutical companies, advocacy groups, and immune and developmental therapeutic programs; and promotes the engagement and involvement of minority participants in clinical research. Finally, the Core is developing a program to involve patient advocacy and support groups to ensure that MSK sarcoma clinical trial protocols best serve patient needs and generally stimulates conversation between researchers and patients, caregivers, and advocates.
Developmental Research Program
Program Directors:
Marc Ladanyi, MD, Director
Samuel Singer, MD, Co-Director
To enable SPORE investigators to rapidly develop innovative research programs with strong potential for clinical impact, the Developmental Research Program (DRP) provides seed funding for pilot projects on the pathogenesis, progression, and treatment-driven evolution of sarcoma. With institutional support, the MSK SPORE in Soft Tissue Sarcoma DRP provides up to 4 awards annually. Projects that yield promising data will be considered for incorporation into the SPORE as Research Projects in future funding cycles.
Career Enhancement Program
Program Directors:
Meera R. Hameed, MD, Director
Hedvig Hricak, MD, PhD, Co-Director
The Career Enhancement Program (CEP) encourages more translational researchers to focus on soft tissue sarcoma research by providing research support for early-stage projects proposed by junior investigators. One to two awardees per year are selected based on their previous accomplishments and their commitment to a career in academic sarcoma research. The CEP also enhances awardees’ career training by providing sarcoma-specific mentorship and integration into the SPORE program through participation in conferences and presentations.







