SPORE in Prostate Cancer
Johns Hopkins University
Principal Investigator(s):

Samuel Denmeade, MD

Shawn Lupold, Ph.D.
- Principal Investigator(s) Contact Information
- Overview
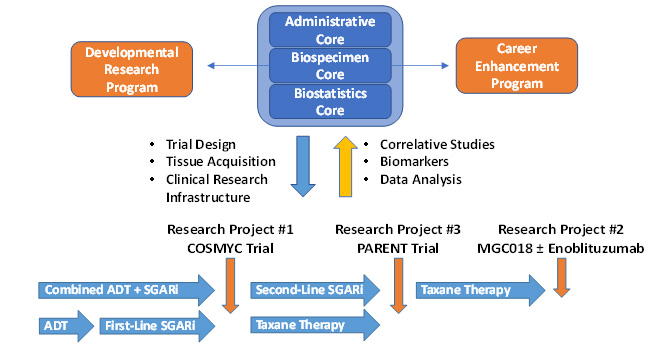
- Project 1: Combined Suppression of MYC as Therapy for CRPC; the COSMYC Trial
- Project 2: Dual Targeting of B7-H3 in Metastatic Prostate Cancer
- Project 3: Induction of Synthetic Lethality with Combined DNMT and PARP Inhibition
- Administrative Core
- Biospecimen and Pathology Core
- Biostatistics and Bioinformatics Core
- Developmental Research Program
- Career Enhancement Program
Principal Investigator(s) Contact Information
Samuel Denmeade, MD
R. Dale Hughes Professor of Oncology
201 North Broadway, Viragh 9121, Box 7
Baltimore, Maryland, 21287-0031
(410) 955-8875 (desk)
(410) 733-3232
Shawn Lupold, Ph.D.
Professor, Johns Hopkins University
600 N. Wofle St. Park 209
Baltimore, Maryland 21287
(410) 502-4822
Overview
Prostate cancer (PCa) is the most frequently diagnosed cancer in men in the United States. More than 30,000 American men die from PCa each year, and despite considerable progress over the last decade, the cure of metastatic prostate cancer has remained elusive. New research has identified the continued addiction to androgen receptor signaling and a tumor environment that suppresses the patients immune system as a fundamental characteristic of PCa that produce resistance to hormonal and immune-based therapies. Genomic studies revealed the magnitude of DNA repair mutations in men with prostate cancer leading to the approval of PARP inhibitors in a subset of men with select mutations in these pathways. To make significant advancement in the treatment of prostate cancer, the goal of this Johns Hopkins Prostate Cancer (JHPC) SPORE is to use new insights to develop innovative combination therapies in clinical trials. The overall objective of the JHPC SPORE is to reduce death and suffering from prostate cancer mortality by developing better therapies for the disease. JHPC SPORE investigators have generated three Research Projects that evaluate innovative hormonal, DNA-targeted and immune-based combination treatment strategies. These SPORE Investigators from five different Departments make up a highly collaborative team that will share expertise, reagents, techniques, equipment, and infrastructure across all three Research Projects. They will meet frequently together to review progress. The Overall Aims of this SPORE application are 1) to evaluate the safety and efficacy of new hormonal, DNA-targeted and immunotherapeutic combination therapy strategies for the treatment of prostate cancer in three separate clinical trials; 2) to assess the ability of these new modalities to overcome resistance pathways; 3) to develop cancer models to determine the effectiveness of additional treatments with the goal of identifying effective combination strategies to eliminate prostate cancer. These projects are supported by three Core Resources, two Developmental Research Projects, and two Career Enhancement Projects, that have been chosen to take advantage of the most recent advances in the understanding of PCa molecular and immune biology in order to speed up the development of innovative new treatments.
Research Structure of the Johns Hopkins Prostate Cancer SPORE
Project 1: Combined Suppression of MYC as Therapy for CRPC; the COSMYC Trial
Project Co-Leaders:
Samuel Denmeade, MD (Clinical Co-Leader)
Angelo De Marzo, MD, Ph.D. (Basic Science Co-Leader)
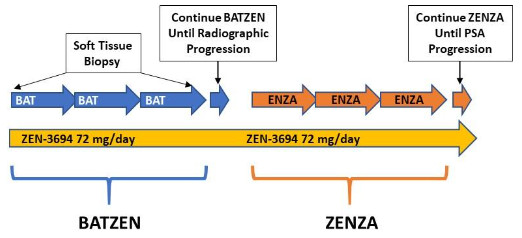
Prostate cancer cells (PCa) are able to adjust to changes in their environment, especially in response to low testosterone levels caused by Androgen Deprivation therapy (ADT). Long-term exposure to ADT causes PCa to greatly increase the number of androgen receptors (AR) to try to restore hormone signaling required for cancer growth. This adaptive increase in AR makes them more resistant to further hormone therapy. Our research shows that ADT-resistant cells with high AR levels can paradoxically be killed by high levels of testosterone. The testosterone causes surviving PCa to decrease the amount of AR, making them once again sensitive to AR-blocking treatments like enzalutamide.
We called this new approach Bipolar Androgen Therapy (BAT). Clinical trials with BAT have shown that it safe for men with asymptomatic metastatic castrate-resistant PCa. Across studies, about 30% of patients experienced a disease response. BAT improved sexual function, fatigue and physical activity. BAT also restored sensitivity to subsequent treatment with enzalutamide.
These trials established BAT as a new type of treatment for PCa. The goal of this project is to try to make the effect of BAT last longer. We found that BAT can block the production of a protein called MYC. MYC is a powerful growth signal that gets turned on in hormone-resistant PCa. A class of drugs known as BET bromodomain inhibitors can also turn off the MYC protein. We have found that one of these BET inhibitors, ZEN-3694, can enhance the effectiveness of BAT to kill PCa cells.
Specific Aims:
Aim 1: Perform the COSMYC clinical trial in men with castrate resistant prostate cancer to test the effectiveness and safety of BAT and ZEN-3694 in sequence with enzalutamide as a new combination therapy.
Aim 2: Evaluate the effects of BAT and ZEN-3694 on the growth and death signals in PCa cells using biopsies from patients on the trial
Aim 3: Explore whether DNA in the blood of treated patients can be used to predict who will respond to the therapy and when to switch from one therapy, BAT, to the other therapy, enzalutamide.
The COSMYC trial:
Project 2: Dual Targeting of B7-H3 in Metastatic Prostate Cancer
Project Co-Leaders:
Kenneth J. Pienta, MD (Clinical Co-Leader)
Drew M. Pardoll, MD, Ph.D. (Basic Co-Leader)
The overall goal is to provide the first in-human assessment of clinical activity in relation to targeted treatment against B7-H3 mediated by cytotoxic antibody drug conjugate (ADC) +/- immunological antibody dependent cellular cytotoxicity (ADCC) activity to determine if synergistic anti-tumor activity is observed.
Specific Aims:
Aim 1: We will conduct an investigator-initiated randomized (1:1) phase II clinical trial utilizing an anti-B7-H3 ADC and anti-B7-H3 ADCC monoclonal antibody in men with metastatic prostate cancer (mPC) to assess clinical benefit based on overall response rate (ORR). Key secondary endpoint will be duration of response (DoR), PSA50 response, progression free survival (PFS), overall survival (OS), and correlative (pharmacodynamic, immunologic) endpoints will be included to allow interrogation of mechanisms underlying the activity of low-dose ADC and whether it can be further potentiated by concomitant T cell and myeloid cell enhancement observed with the ADCC monoclonal antibody.
Aim 2: We will assess and quantify tumor cell proliferation and apoptosis following single and dual B7-H3-targeted treatment. In matched pre- versus post-treatment biopsy specimens, we will employ single and multi-parameter IHC to quantify measures of intratumoral T cell response, assess myeloid and NK cell densities and functional status, and assess adaptive therapy-induced expression of the targetable immune checkpoints.
Aim 3: We will interrogate functional antigen-specific T cell repertoires induced by single or dual targeting of B7-H3 in mPC. This will allow exploration of the breadth and functional state of tumor-specific T cell responses after single and dual B7-H3-targeted therapies in patients with mPC.
The overall translational impact of these studies includes:
- Understanding if combining ADC with ADCC agents at lower doses allows improved tolerability and efficacy compared to monotherapy at higher doses.
- Integrate novel spatial proteomic/transcriptomic, T cell repertoires, and functional analyses in prostate cancer patients thereby potentially identifying critical immune molecules and pathways that can be targeted for future combinatorial immunotherapy prostate cancer trials.
Project 3: Induction of Synthetic Lethality with Combined DNMT and PARP Inhibition
Project Co-Leaders:
Srinivasan Yegnasubramanian, MD, Ph.D. (Basic Co-Leader)
Michael Carducci, MD (Clinical Co-Leader)
Metastatic prostate cancer can be aggressive, especially when it resists standard therapies. Recent research has highlighted the effectiveness of class of drugs called PARP inhibitors in treating metastatic castration-resistant prostate cancer (mCRPC). These inhibitors work particularly well in patients with specific genetic alterations related to homologous recombination deficiency (HRD), such as BRCA1 or BRCA2 mutations. However, only a small percentage of mCRPC patients have these mutations and benefit from PARP inhibitors.
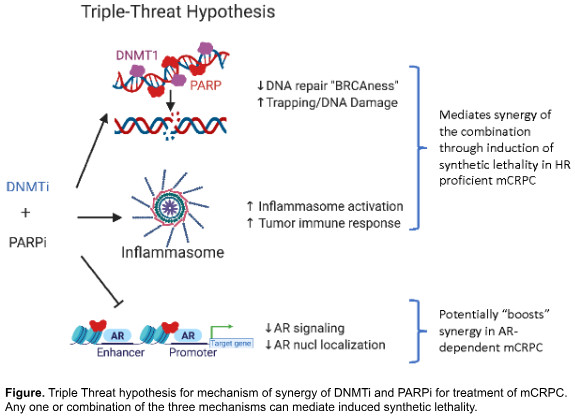
Our innovative approach aims to broaden the impact of PARP inhibitors. We propose combining them with another type of drug called DNA methyltransferase inhibitors (DNMTi). We hypothesize that this combination could be effective in treating prostate cancers even when they don’t have an underlying HRD, by creating a “triple-threat” against prostate cancer cells:
- DNA Repair Suppression: reduces the cancer cells’ ability to repair DNA damage, making them vulnerable.
- Immune Activation: activates immune pathways, promoting anti-tumor immune responses.
- AR Signaling Inhibition: blocks AR signaling, crucial for prostate cancer cell survival, we target mCRPC directly.
Our research involves rigorous testing.
Specific aims:
Aim 1: we will study this combination in lab models and animal tests.
Aim 2: we will confirm the mechanisms by which it induces synthetic lethality in mCRPC.
Aim 3: we’ll conduct a clinical trial with advanced mCRPC patients.
If successful, our work could revolutionize prostate cancer treatment, by developing a new combination treatment for men with prostate cancers that don’t have HRD mutations.
Administrative Core
Core Co-Leaders:
Samuel Denmeade, MD (Clinical Co-Director)
Shawn Lupold, Ph.D. (Basic Science Co-Director)
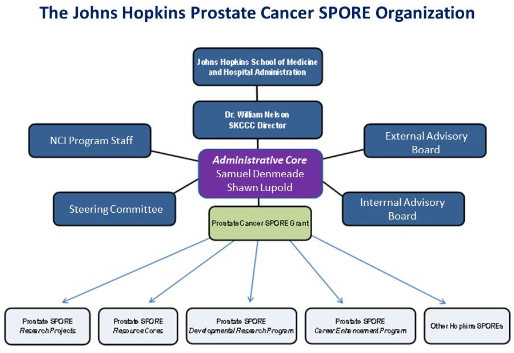
The goal of the Administrative Core is to provide scientific and administrative oversight for all the program activities of the Johns Hopkins Prostate Cancer (JHPC) SPORE. In addition, the Core will facilitate interactions between JHPC SPORE Investigators and investigators associated with other Prostate Cancer initiatives at other Cancer Centers. The managerial structure of JHPC SPORE is designed to promote patient-focused research by creating an inclusive prostate cancer research culture across departments at Johns Hopkins. The core provides high-quality monitoring, evaluation, and oversight of the SPORE portfolio of Research Projects and Core Resources. The core oversights the review and selection of the Career Enhancement Program, and the Developmental Research Program projects with the goal of stimulating talented young investigators to explore new research opportunities. The core will regularly consult with the External and Internal Advisory boards to discuss the progress of research projects and other Core functions. The Core also provides communications, resources, including teleconferences, travel funds, and administrative staffing for all its managerial activities. With strong teams and advocates, and through new and existing collaborations, the Core will ensure that JHPC SPORE investigators continue to provide leadership and transform care in our community, nationally, and internationally.
Biospecimen/Pathology Core
Core Co-Leaders:
Angelo DeMarzo, MD, Ph.D. (Clinical Co-Director)
Tamara Lotan, MD (Basic Co-Director)
Core B will maintain and enhance a repository of prostate tissues, provide expert diagnoses of tissue specimens, and continue to develop state-of-the-art protocols for tissue-based technology. It will also aid in the interpretation and quantitative image analyses of various staining methods, design and distribute tissue microarrays, and implement best practices for tissue acquisition, processing, annotation, storage, and distribution. The project will continue the Johns Hopkins Rapid Autopsy program.
The project involves three main studies: the COSMYC trial, which focuses on the suppression of MYC as therapy for CRPC; a study on dual targeting of B7-H3 in metastatic castrate-resistant prostate cancer; and a study on the induction of synthetic lethality with combined DNMT and PARP inhibition. These studies will involve the collection and processing of tissue biopsies, the performance of various assays, and the development of correlative studies. The project aims to advance our understanding of prostate cancer and contribute to the development of more effective treatments.
Biostatistics/Bioinformatics Core
Core Co-Leaders:
Hao Wang, Ph.D. (Clinical Co-Director)
Bruce Trock, Ph.D. (Basic Co-Director)
The Biostatistics and Bioinformatics Core provides state-of-the-art biostatistics and bioinformatics support to ensure the scientific rigor and reproducibility of all results in the Prostate Cancer SPORE. The Core participates in study design, data collection, database development and management, data quality control, clinical trial monitoring, data analysis and visualization, bioinformatics analysis, interpretation, and data sharing. As the central hub for quantitative research expertise, the Core supports SPORE investigators, including developmental research program and career enhancement program awardees, as well as other cores. Core members are integral team players in each project, offering solutions to both routine and complexed or unique problems encountered during the project planning and execution. The core also researches methods for statistical and bioinformatics problems that don’t have standard methods or software. Each project is assigned a primary statistician with specialized knowledge in the relevant subject matter, ensuring effective and tailored support. Additionally, the collective expertise of the entire Core team is available to all projects, fostering a collaborative and innovative research environment for the SPORE.
Developmental Research Program
Program Directors:
Jun Luo, Ph.D. (Co-Director)
Marikki Laiho, Ph.D. (Co-Director)
The Developmental Research Program (DRP) is a special feature of the SPORE grant that allows sponsored institutions to fund important new pilot projects with promising translational potential, to generate sufficient preliminary data for an independently funded NIH grant. This is a crucial part of our SPORE as it provides a pipeline and testing ground for novel substrates for translational impact. These projects are intended to last one year with the possibility of a second year of support with the demonstration of sufficient progress and are budgeted with the intention that the majority of the funds be spent on supplies, rather than salary support. Pilot projects that make significant progress and are deemed to be capable of high translational impact will be considered for promotion to full SPORE projects. To be eligible, the applicant must have a current academic appointment at any of the Johns Hopkins Medical Institutions. Applicants must hold an M.D., or Ph.D. degree or both. The applicants are expected to provide evidence of a significant research commitment to ensure that the proposal can in fact be addressed in an effective and productive way. The Developmental Research Program will be maintained throughout the grant period.
Career Enhancement Program
Program Directors:
Elizabeth Platz, Sc.D. (Co-Director)
Otis Brawley, M.D. (Co-Director)
The Career Enhancement Program (CEP) is a special feature of the SPORE grant which allows sponsored institutions to support junior faculty or established investigators who wish to enhance or refocus their careers on translational research in the field of prostate cancer. This is a crucial part of our SPORE as it provides funding to promising young investigators to facilitate their early career development and to evolve into independent investigators. It also can supply funds to more senior investigators who have not previously focused on prostate cancer and who wish to direct their efforts to the area of translational prostate cancer research. These projects are intended to last for two years with the second year of funding dependent upon the supported investigator making progress towards the stated aims and goals of the project. To be eligible, the applicant must have a current academic appointment at any of the Johns Hopkins Medical Institutions. Applicants must hold an M.D., or Ph.D. degree or both. The applicants are expected to provide evidence of a significant research commitment to ensure that the proposal can in fact be addressed in an effective and productive way.







