Dana-Farber Harvard Cancer Center (DF/HCC) SPORE in Prostate Cancer
Dana-Farber Harvard Cancer Center
Principal Investigator(s):

Himisha Beltran, MD

Steven Balk, MD, PhD
- Principal Investigator(s) Contact Information
- Overview
- Project 1: Efficacy and Mechanisms of Resistance to Neoadjuvant Intensive Androgen Signaling Inhibition
- Project 2: Molecular Determinants of Response and Resistance to EZH2 and PARP inhibition in Prostate Cancer
- Project 3: Dissecting and Predicting Lethal Prostate Cancer using Biologically Informed Artificial Intelligence
- Administrative Core
- Biospecimen Core
- Biostatistics/Computational Core
- Developmental Research Program
- Career Enhancement Program
Principal Investigator(s) Contact Information
Himisha Beltran, MD
Associate Professor of Medicine in the Lank Center for Genitourinary Oncology
Director of Translational Research within Medical Oncology
Dana-Farber Cancer Institute
450 Brookline Avenue, Dana 1420
Boston, Massachusetts, 02215
(617) 582-9421
Steven Balk, MD, PhD
Professor of Medicine
Beth Israel Deaconess Medical Center
3 Blackfan Circle, 443 CLS
Boston, MA 02115
(617) 735-2065
Overview
The Dana-Farber/Harvard Cancer Center (DF/HCC) Prostate Cancer SPORE seeks to improve the understanding and treatment of prostate cancer using a highly translational approach. The SPORE infrastructure will facilitate interactions and collaboration within our thriving community of basic, clinical, and population science researchers dedicated to prostate cancer research. Each project addresses a fundamental challenge that contributes to prostate cancer morbidity and mortality.
Project 1 leverages tumor specimens from patients with high-risk prostate cancer treated with neoadjuvant therapies to understand how tumors respond and resist acute potent androgen receptor blockade and to develop novel strategies to improve cure rates and combat resistance.
Project 2 will develop innovative strategies to target the epigenome in later stages of advanced castration resistant prostate cancer and will develop a first-in-field clinical trial focused on co-targeting EZH2 and PARP.
Project 3 delves deep into biomarkers in localized prostate cancer, leveraging innovations in computation and biologically guided deep learning, to deliver on precision cancer medicine in this disease state. The ability to understand why some localized prostate cancers are phenotypically aggressive and predict which localized prostate cancers will behave in this manner addresses a large clinical unmet need.
Each of these projects combines elegant preclinical work with innovative clinical studies led by DF/HCC investigators. The Administrative Core is the center for scientific, fiscal, and administrative oversight. The Biostatistics and Computational Biology core will provide specialized expertise in biostatistics and the management of genomic and other next generation sequencing data and data sharing. The Biospecimen Core will maintain tissue/blood repositories for the SPORE projects as well as other investigators within the prostate cancer program. The Developmental Research and Career Enhancement Programs will identify and fund innovative projects that address basic, translational, and clinical research questions and unmet needs in prostate cancer and will support early career and new prostate cancer investigators. We anticipate that the DF/HCC Prostate SPORE will make substantial scientific discoveries in the field and translate directly into benefits for men with prostate cancer.
Project 1: Efficacy and Mechanisms of Resistance to Neoadjuvant Intensive Androgen Signaling Inhibition
Project Co-Leaders:
Steven Balk, MD, PhD (Basic)
Mary Ellen Taplin, MD (Clinical)
Neoadjuvant trials provide a valuable opportunity to increase cure rates for men with high-risk prostate cancer as well as our understanding of response and resistance to systemic therapies. We hypothesize that intensive neoadjuvant androgen signaling inhibition (ASI) therapy, in addition to decreasing tumor burden, will translate into an improvement in disease free survival and ultimately overall survival. We have been testing this hypothesis in a series of phase 2 trials examining the efficacy of neoadjuvant intensive ASI using leuprolide in combination with abiraterone (ABI) and/or enzalutamide (ENZ) or apalutamide (APA) for 6 months prior to radical prostatectomy. Responses appear to be improved relative to previous neoadjuvant trials, but it remains unclear whether the cases with pathologic complete response (pCR) or minimal residual disease (MRD) after therapy reflect tumors that have less metastatic potential. Moreover, the molecular basis for residual disease in prostate, and its relationship to metastatic disease in patients who progress, remain to be determined.
Specific Aims
Aim 1: Address the molecular basis of response versus resistance, we propose comprehensive genomic analyses of tumors with exceptional responses (pCR/MRD) to intensive neoadjuvant ASI therapy, and of residual disease in prostatectomy specimens in men who do not achieve pCR/MRD.
Aim 2: Identify actionable acute nongenomic adaptations that mediate initial resistance to intensive ASI therapy, and specifically test the hypothesis that these adaptations converge on expression of D cyclins and activation of CDK4/6 to drive proliferation.
Aim 3: Perform a randomized phase 2 trial of intensive ASI (leuprolide plus dirlotapides), alone or in combination with the CDK4/6 inhibitor abemaciclib. Our long-term goal is to establish neoadjuvant trials as a platform for assessing the efficacy of novel combination therapies in hormone sensitive prostate cancer.
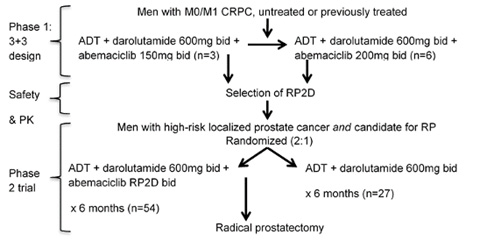
Figure: Trial Schema. In Phase 1, a 3+3 design will be used to define the recommended phase 2 dose (RP2D) of abemaciclib. The starting dose will be abemaciclib 150mg bid with the aim of escalating this to 200mg bid if tolerated. A total of 3-12 patients are anticipated in phase 1, and they will continue on therapy until radiographic progression or investigator determination of lack of ongoing benefit. In Phase 2, 81 men with high-risk localized disease will be randomized 2:1 to 6 months of LHRHa +darolutamide + abemaciclib (Arm 1) or LHRHa + darolutamide.
Project 2: Molecular Determinants of Response and Resistance to EZH2 and PARP inhibition in Prostate Cancer
Project Co-Leaders:
Himisha Beltran, MD (Clinical)
Myles Brown, MD (Basic)
Epigenetic dysregulation, including overexpression of the enhancer of zeste homolog 2 (EZH2), drives treatment resistance in prostate cancer. Beyond its canonical role as a member of the PRC2 complex as a transcriptional repressor, we previously identified a non-canonical role of EZH2 in activating AR signaling, as well as in driving lineage plasticity and neuroendocrine prostate cancer. Current data suggests that single agent activity may be limited and that combination strategies should be pursued. We discovered a novel interaction between EZH2 and DNA repair processes and a synergy between EZH2 and PARP inhibition in prostate cancer. We hypothesize that EZH2 drives downstream molecular programs through canonical and non-canonical mechanisms, and that these downstream effects are context dependent. Combination therapy with EZH2 and PARP inhibition will therefore have differential biologic impact in prostate cancer based on the underlying genomic, epigenomic, and phenotypic context. We will elucidate the mechanisms by which EZH2 regulates DNA repair and define how DNA repair pathways impact EZH2 drug sensitivity. We will evaluate the biologic and clinical impact of EZH2 and PARP inhibition across the heterogeneous spectrum of advanced prostate cancer through in-depth preclinical and clinical studies. We will conduct a first-in-field, investigator-initiated clinical trial of the EZH2 inhibitor tazemetostat plus PARP inhibitor talazoparib with extensive translational correlates. Results from this project will provide the basis for a new therapeutic strategy for men with advanced prostate cancer.
Specific Aims:
Aim 1: Elucidate the crosstalk between EZH2 and DNA repair pathways and mechanisms of synthetic lethality.
Aim 2: Define the biologic impact of EZH2 + PARP inhibition across the heterogeneous spectrum of CRPC including both AR-driven and non-AR-driven tumors.
Aim 3: Identify biomarkers of response and resistance to the combination of EZH2 + PARP inhibition in patients.
Project 3: Dissecting and Predicting Lethal Prostate Cancer using Biologically Informed Artificial Intelligence
Project Co-Leaders:
Eliezer Van Allen, MD (Basic)
Chin-Lee Wu, MD, PhD (Clinical)
Multiple molecular factors, including germline and somatic alterations in DNA repair genes and tissue-based transcriptional biomarkers, have biological and prognostic relevance in prostate cancer. Determination of the interacting and co-occurring molecular features that jointly drive indolent or aggressive clinical outcomes is urgently needed to enable molecularly guided therapeutic strategies and biologically grounded predictive models for clinical use. Furthermore, complex molecular states may converge on histopathological patterns to augment these predictions, but these properties are difficult to quantify, integrate, and generalize across diverse patient populations. The advent of large and diverse patient cohorts with clinically embedded molecular characterization, digital histopathology techniques, and key outcome measures, along with innovations in computation and deep learning to analyze and interpret these data, has created an opportunity to profoundly expand the discovery and translational potential of molecular, pathologic, and phenotypic data for patients with localized prostate cancer. Our overarching hypothesis is that interacting molecular, pathologic, and phenotypic features define prognostic outcomes in intermediate and high-risk localized prostate cancer, and that biologically guided interpretable deep learning, paired with harmonized cohorts representative of prostate cancer diversity, will transform our understanding of indolent versus potentially lethal disease and deliver on the promise of precision cancer medicine.
Specific Aims:
Aim 1: Dissect the interacting germline and somatic properties using biologically guided neural networks.
Aim 2: Determine the convergent spatial histopathologic properties of molecularly and clinically distinct forms of prostate cancer.
Aim 3: Develop and validate a clinical grade molecular prognostic model guided by biological networks in real-world and clinical trial settings. This project strives to transform precision cancer medicine for prostate cancer and serve as a model for the creation, development, and application of these emerging methodologies across cancer types and contexts.
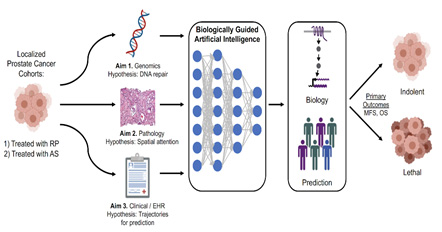
Figure: Overarching schematic of this proposal
Administrative Core
Core Co-Directors:
Himisha Beltran, MD
Steven Balk, MD, PhD
The DF/HCC Prostate SPORE Administrative Core (Core A) is instrumental for the overall function and success of the Prostate SPORE program. The Directors will work closely with Advisory Board Members, Patient Advocates, and SPORE investigators to foster an environment that promotes collaboration and transparency in research. Regular reviews and fiscal oversight by Core A will ensure that the program has the resources it needs to be successful. Core A will also ensure that the SPORE integrates well with the broader DF/HCC program and is capable of leveraging resources and institutional infrastructure to accelerate translational prostate cancer research.
Specific Aims
Aim 1: Provide resources and fiscal oversight.
Aim 2: Monitor research progress and plan for the future.
Aim 3: Integrate the SPORE within the Dana Farber/Harvard Cancer Center.
Aim 4: Foster collaborative research and dissemination of research findings.
Aim 5: Integrate a patient perspective throughout all research activities in the Prostate SPORE.
Biospecimen Core
Core Co-Directors:
Sabina Signoretti, MD
Chin-Lee Wu, MD, PhD
Acquisition and characterization of biospecimens linked with clinical data and the development of patient- relevant preclinical models, are essential components for effective and impactful translational research. The Dana Farber/ Harvard Cancer Center (DF/HCC) Prostate SPORE Biospecimen Core (Core C) will maintain and expand a mature prostate cancer biorepository, including blood and tissue specimens, from a diverse group of prostate cancer patients across DF/HCC who have provided consent to our clinical database and biobank protocols. Core C will provide tissue specimens and vast pathology expertise to SPORE investigators including the evaluation of both human and mouse model tissues. Core C will maintain a database of clinical data on all consented patients, linked with standardized pathology review procedures and data collection. Core C will work closely with SPORE investigators and collaborators to provide interpretative and consultative pathology services, including state-of-the-art molecular assays and imaging technologies developed at DF/HCC. Centralization of Core activities builds upon established infrastructure but does not replicate existing resources within the DF/HCC. Overall, Core C will greatly facilitate and accelerate translational research within the DF/HCC Prostate SPORE through the collection and integration of highly annotated biological material and associated clinical information.
Specific Aims
Aim 1: To provide a specimen bank for SPORE.
Aim 2: To maintain a database of key clinical variables.
Aim 3: To provide pathology services to SPORE investigators.
Biostatistics/Computational Core
Core Director:
Svitlana Tyekucheva, PhD
The Biostatistics and Computational Core (Core B) interacts with all research activities within the SPORE to ensure the highest standards of scientific rigor in areas of study design, data management and integrity, and data analysis and interpretation. The overarching goal is to promote translational research derived from fundamental discoveries in the laboratory that can lead to tangible clinical benefit.
Specific Aims
Aim 1: Provide biostatistical and computational biology expertise for the planning and design, conduct, analysis, and reporting of laboratory, genomic, animal, translational, clinical, and epidemiological studies.
Aim 2: Provide consultation on issues of data management, integrity and integration, including data collection, storage, transfer, sharing and quality assurance, on statistical and computational software and programs, and on coordination of laboratory results with parameters and outcomes from clinical studies.
Aim 3: Provide short-term biostatistics and computational biology consulting to SPORE researchers.
Core B has a wealth of experienced biostatisticians and computational biologists equipped with excellent computational resources, including major commercial and public-use statistical software and a large library of locally written software for design and analysis.
Developmental Research Program
Program Co-Directors:
Matthew Freedman, MD
Mark Pomerantz, MD
The objective of the Developmental Research Program (DRP) is to identify and support exciting prostate cancer research and ensure a continuous renewal of high-quality scientific endeavors within the SPORE. The program will recruit diverse disciplines and backgrounds to bring new approaches to prostate cancer. Applications will be solicited broadly and reviewed fairly by a highly qualified and engaged committee. DRP recipients will become part of the SPORE community with exposure, feedback, and access to core resources. Overall, the DRP will maximize the chance that new and cutting-edge concepts supported by promising pilot data have the capability to receive larger grant funding and ultimately impact the field.
Specific Aims
Aim 1: Foster the development of innovative and promising prostate cancer research by providing seed funding.
Aim 2: Identify and prioritize solicitations focused on areas with potential to have the highest impact of improving the outcome of patients.
Aim 3: Monitor the progress of developmental projects, provide critical and timely feedback, and encourage and enable promising DRP projects to compete for extramural peer- reviewed funding.
Aim 4: Bring new investigators into prostate cancer translational research.
Career Enhancement Program
Program Directors:
Adam Feldman, MD, MPH
Lorelei Mucci, ScD
Paul Nguyen, MD
The goal of the Career Enhancement Program (CEP) is to provide resources, financial support, and mentorship for investigators who are transitioning or have recently transitioned to independent positions. The CEP will attract and nurture careers in clinical, basic, and population science. In turn, the DF/HCC Prostate SPORE program will greatly benefit through interactions with fresh and diverse ideas and talent. Applications will be solicited broadly across DF/HCC and Massachusetts and will undergo a fair and transparent review process. The CEP program will help advance the research careers of faculty by providing funding for independent projects, infrastructure and resources through the cores, interactions with established prostate cancer investigators through SPORE meetings and educational sessions, and by establishing a formalized mentorship program. The CEP program will build upon a strong prior track record of success in enhancing and supporting career development and is a critical piece of the overall SPORE mission to train and develop the next generation of leaders in prostate cancer translational research.
Specific Aims
Aim 1: Develop a transparent and supportive system to identify and nurture promising early-career investigators seeking to become independent investigators in translational prostate cancer research.
Aim 2: Provide funding, resources, career development, and ongoing support for awardees’ innovative prostate cancer research projects; monitor their research progress; facilitate collaborations to enhance clinical translation; and provide opportunities for sustaining these individuals in prostate cancer.
Aim 3: Establish a formalized mentoring program and promote a supportive environment through SPORE related activities that will assist in the career development of the award recipient and other early career members.
Aim 4: Develop a pipeline of the next generation of leaders in prostate cancer translational research, and attract and retain women and underrepresented racial and ethnic groups who will make critical contributions to translational prostate cancer research.







