SPORE in Skin Cancer
Wistar Institute
Principal Investigator(s):

Meenhard Herlyn, DVM, DSc

Ravi Amaravadi, MD
- Principal Investigator(s) Contact Information
- Overview
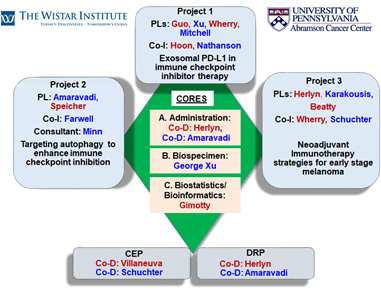
- Project 1: Exosomal PD-L1 in immunotherapy resistance
- Project 2: Targeting autophagy to improve melanoma immunotherapy
- Project 3: Immunotherapy strategies for early-stage melanoma
- Administrative Core
- Biospecimen and Pathology Core
- Biostatistics & Bioinformatics Core
- Developmental Research Program
- Career Enhancement Program
Principal Investigator(s) Contact Information
Meenhard Herlyn, DVM, DSc
Professor; Director, Melanoma Research Program
Wistar Institute
3601 Spruce Street
Philadelphia, PA 19104
(215) 898-3950
Ravi Amaravadi, MD
Co-Leader of the Cancer Therapeutics Program, Abramson Cancer Center; Associate Professor of Medicine, Hospital of the University of Pennsylvania
University of Pennsylvania
Abramson Cancer Center, West Pavilion, 3rd Floor, 3400 Civic Center Boulevard
Philadelphia, PA 19104
Overview
The Wistar/University of Pennsylvania SPORE in Skin Cancer is the result of a decades-long collaboration between the two institutions in the effort to reduce the morbidity and mortality from skin cancer. The clinical landscape of melanoma has evolved rapidly over the last decade. Immune checkpoint inhibition has revolutionized melanoma therapy to the point where every high-risk melanoma patient will be treated at some point with these agents. However, many major questions remain on how best to use these immune therapeutics. This SPORE has three melanoma-focused projects:
Project 1: Exosomal PD-L1 in immune checkpoint inhibitor therapy. Project 1 addresses the unmet need to find an effective biomarker to select patients for single agent versus combination immunotherapy. Exosomal PD-L1 is an immunosuppressive factor secreted by melanomas. We propose rigorous clinical utility studies designed to demonstrate this blood-based measurement as a highly-sensitive and specific predictive biomarker for anti-PD-1 antibody (Ab)-based therapy.
Project 2: Targeting autophagy to improve immune checkpoint inhibition. Project 2 addresses a second unmet need for a safer and effective combination regimen that promises to be effective in anti-PD-1 Ab refractory patients. Based on extensive preclinical data and a new molecular target in the lysosome, we have developed a clinical trial of combined anti-PD1 Ab or anti-PD-1 and anti-CTLA-4 Ab and lysosomal autophagy inhibition, a new strategy for reprogramming tumor-associated macrophages to enhance the efficacy of T-cell killing.
Project 3: Neoadjuvant immunotherapy strategies for early-stage melanoma. Project 3 fills a major gap in the treatment of early disease by conducting a clinical trial with anti-PD1 Ab in stage IIB/C melanoma patients. Besides in-depth characterization of the immune response, the project’s preclinical studies will lead to new strategies for enhancing the immune stimulatory capacity of dendritic cells in the tumor microenvironment.
In addition to these 3 projects this SPORE has a well-developed Developmental Research Project Program and Career Enhancement Project Program. These pilot programs feed the sustainability of the SPORE and will have a special focus on non-melanoma skin cancers and the recruitment of underrepresented minorities into skin cancer research and skin cancer clinical trials.
The highly translational projects and pilot programs are supported by three cores:
Core A: Administrative Core
Core B: Biospecimen and Pathology Core
Core C: Biostatistics and Bioinformatics Core
Project 1: Exosomal PD-L1 in immunotherapy resistance
Project Co-Leaders:
Wei Guo, PhD (Basic)
Xiaowei Xu, MD, PhD (Clinical)
Tara Mitchell, MD (Clinical)
E. John Wherry, PhD (Basic)
Immune checkpoint inhibitors (ICI) such as anti-PD-1 antibodies have revolutionized anti-tumor therapy for many types of cancers including metastatic melanoma. However, patient response rates are low. Combined therapies such as ipilimumab and nivolumab produce a higher response rate but are associated with significant toxicities. A major unmet need is to develop quantitative assays that stratify patients who will respond to anti-PD-1 therapy to avoid unnecessary toxicities, and direct non-responders to alternative treatments. A pre-treatment or early on-treatment predictive biomarker would provide decision-enabling information to clinicians to optimize treatment of melanoma patients. Exosomes are nano-sized vesicles secreted by cells to the extracellular milieu. We found that metastatic melanoma cells secrete exosomes enriched with PD-L1, which suppress the function of CD8+ T cells in circulation and facilitate tumor growth. In patients’ plasma, the level of circulating exosomal PD-L1 and its change during the course of anti-PD-1 treatment are associated with patient response to anti-PD-1 therapy (Chen et al., Nature 2018). We also found that, myeloid cells such as macrophages also secrete exosomes that carry PD-L1, which can be selectively and quantitatively measured in patient blood. The overarching goals of Project 1 are to develop a quantitative liquid biopsy-based tool that enables clinicians to predict patient response to ICI-based therapies, and to understand the role of macrophage-derived exosomes in immune suppression. In Aim 1, we will test the hypothesis that exosomal PD-L1 are effective predictors of patient response to ICI. We perform the assays using a large multi-institutional validation set of patient samples across different major therapeutic contexts, taking advantage of the unique infrastructure SPORE offers.
In Aim 2, we will systematically investigate the pivotal roles of TAM-derived exosomes in immune suppression. The proposed study will also unveil a role of TAM exosomes in immune suppression, which will not only advance our understanding of immune suppression at new dimensions, but also helps develop novel therapeutic approaches to improve the treatment of patients with melanoma.
Project 2: Targeting autophagy to improve melanoma immunotherapy
Project Co-Leaders:
Ravi Amaravadi, MD (Clinical)
David Speicher, PhD (Basic)
New approaches that can overcome therapy resistance to immune checkpoint inhibition (ICI) are a major unmet need for Stage IV melanoma patients. This project will test our overall hypothesis that lysosomal autophagy inhibition results in very focused cellular pathway perturbations in cancer cells and immune cells that enhance the efficacy of ICI.
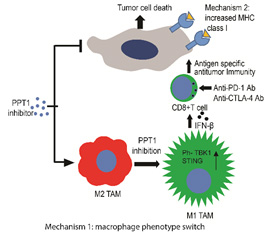
Autophagy has been identified as a major resistance mechanism to chemotherapy and targeted therapy. In a previous SPORE cycle, we combined BRAF/MEK inhibition and the lysosomal autophagy inhibitor hydroxychloroquine (HCQ) and found a high response rate in BRAF mutant melanoma (NCT02257424). The safety and activity of this regimen led to the launch of EA6191 a national randomized trial (NCT04527549). Previously, our group made the discovery that palmitoyl protein thioesterase 1 (PPT1), a lysosomal protein that regulates autophagy, is the primary target of chloroquine derivatives (Rebecca et al., Cancer Discovery 2017). We have also generated a series of potent and specific PPT1 inhibitors exemplified by DC661 (Rebecca et al., Cancer Discovery 2019). Recently, we found that combined anti-PD-1 antibody and HCQ produces striking antitumor efficacy in ICI-refractory mouse models of melanoma (Sharma et al., JCI Insight 2020). We found that lysosomal inhibition elicited a phenotype switch in macrophages that enabled T cell immunity and augmented ICI efficacy.
SPORE Project 2 will use next generation chemical autophagy inhibitors that are headed to the clinic to determine the effects of inhibiting different autophagy targets on improving the efficacy of ICI. We will use a novel genetic mouse model of autophagy inhibition that will determine if autophagy in the tumor cell or the immune cell is the primary driver of resistance. Global unbiased multi-omics techniques will be used to study response and resistance mechanisms to this approach in pre-clinical and clinical samples. For the latter we will leverage our currently accruing trial of ICI and HCQ (the LIMIT melanoma trial [NCT04464759]), which includes a novel CD8+ T cell PET scan protocol developed by Dr. Michael Farwell (Radiology—UPENN) to track immune cell infiltration into treated tumors.
Project 3: Immunotherapy strategies for early-stage melanoma
Project Co-Leaders:
Giorgos Karakousis, MD (Clinical Co-Leader)
Gregory Beatty, MD, PhD (Basic Co-Leader)
Meenhard Herlyn, DVM, DSc (Basic Co-Leader)
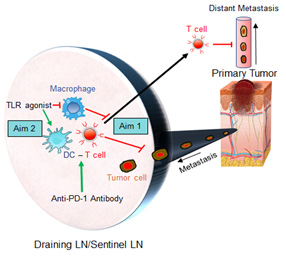
The vast majority of melanomas present with clinically localized disease (clinical stages I and II), contributing significantly to the overall disease burden of this lethal skin cancer. Despite the tremendous therapeutic progress in advanced stage disease, there are limited treatment options for patients with high-risk localized disease. In this project we build upon our success with neoadjuvant anti-PD-1 blockade treatment for patients with clinical stage III disease and will study this approach in a novel clinical trial for patients with clinical stage IIB/C melanoma. In this study, we will investigate the immune microenvironment of the sentinel lymph node (SLN) which may serve as a gatekeeper to prevent lymphatic spread of melanoma but also as an instructor of T cell immunity. Our central hypothesis is that the capacity of the SLN to protect against locoregional and distant melanoma spread is dependent on its immune health which is pliable and determined by the immunostimulatory capacity of lymph node-resident myeloid cells. PD-1 blockade can reinvigorate exhausted tumor-infiltrating T cells; however, many patients fail to respond. We propose that the immunostimulatory capacity of SLN resident dendritic cells (DC) may further explain and predict the responsiveness to anti-PD-1 therapy. In addition to detailed analyses of the SLN immune microenvironment in the setting of PD-1 blockade, we will use mouse models, including a novel humanized mouse model, to decipher the role of DCs in directing outcomes to anti-PD1 therapy. We will additionally study the role of myeloid agonists (e.g., toll-like receptor [TLR] agonists) in enhancing the efficacy of anti-PD1 blockade, with the ultimate goal of improving immunotherapeutic approaches for patients with early-stage melanoma.
Administrative Core
Core Directors:
Meenhard Herlyn, DVM, DSc
Ravi Amaravadi, MD
The essential services provided by the Administrative Core include administrative support for all the investigators in each project and core, fiscal management and oversight for all components of the program project, and organization and communication of all SPORE meetings and activities. The overall goal of this Core is efficient stewardship of the SPORE. The roles of the core Co-Directors and administrative staff are to facilitate communication while stimulating scientific and technological interactions. This will be achieved by:
- Establishing an administrative structure to provide support and management for all Projects, Cores, pilot project activities, and advisory boards;
- Fostering collaborative research; and
- Ensuring compliance with regulatory and governmental guidelines.
Overall, The Administrative Core of this SPORE is designed to provide scientific leadership, effective communication, and an administrative support structure to ensure the coordination of all SPORE activities.
Biospecimen and Pathology Core
Core Directors:
Xiaowei Xu, MD, PhD
The Biospecimen and Pathology Core will provide the following services to projects and integration with other cores in this proposal:
- Collect high-quality biospecimens and maintain a functional biospecimen bank;
- Expand and maintain a functional biospecimen and clinical database;
- Select and distribute high-quality biospecimens;
- Provide comprehensive pathology services; and
- Develop innovative methodology to facilitate clinically impactful research.
This Core seeks to improve the lives of skin cancer patients by facilitating novel melanoma therapies and novel translational biomarker validation. This Core serves as a centralized repository to procure, process, distribute, and analyze malignant and normal tissue and associated data in support of translational skin cancer research. The central coordination and analysis of tissues by Core B will maximize time and cost effectiveness. This Core supports all stages of SPORE projects, Career Enhancement Program (CEP) awardees, Developmental Research Program (DRP) projects, as well as Inter-SPORE collaborations.
Biostatistics & Bioinformatics Core
Core Directors:
Phyllis A. Gimotty, PhD
The goal of the Biostatistics & Bioinformatics Core is to collaborate with SPORE investigators to ensure that SPORE-related studies have high quality data management, study designs and analytic plans that provide an optimal foundation for all statistical inferences, and statistical analyses that are presented in the most informative manner. Core personnel will contribute their expertise and experience in biostatistics and bioinformatics in areas identified by the following specific aims, each designed around provision of:
- A robust computational and data management environment for SPORE-related datasets;
- Expertise on both research methodologies to design and implement rigorous laboratory studies, biomarker studies and clinical trials and statistical methodologies critical to each research project’s objectives, including plans for the data preprocessing and analysis for big data discovery studies, testing proposed statistical hypotheses, and the development of statistical models;
- Expertise to conduct evaluations of different methodologies to identify the most effective approach, to modify methods and develop new methods to be used when standard methods are not appropriate, and to conduct discovery studies using archived public databases relevant to the research projects; and
- Expert interpretation of research data, collaboration with project investigators to make scientifically and statistically appropriate statements, and to assist in the preparation of scientific abstracts, presentations, and manuscripts.
The Biostatistics & Bioinformatics Core will be led by Dr. Phyllis A. Gimotty, a Professor of Biostatistics at the University of Pennsylvania. She has served as the Director of the biostatistics cores of the Wistar/Penn SPOREs on Skin Cancer since 2004 and has substantial experience in biostatistical methodology relevant to translational cancer research. Drs. Andrew Kossenkov, PhD (Wistar) and Rosemarie Mick, MS (UPenn) will serve as co-investigators.
Developmental Research Program
Program Directors:
Meenhard Herlyn, DVM, DSc
Ravi Amaravadi, MD
Building on the success of previous SPORE grant cycles, the co-Directors of the overall SPORE grant will serve as Development Research Program (DRP) co-Directors. The goals remain to advance high-quality research, foster new ideas, and move research studies from pilot to project status. A second goal of the DRP is to create opportunities for the scientific development of junior faculty or senior investigators who are interested in transitioning into skin cancer research. Our approach is summarized in 2 specific aims and is enabled by a strong commitment for an exceptional level of institutional support from The Wistar Institute and University of Pennsylvania. The DRP will, in particular, welcome projects on non-melanoma skin cancers and topics not covered by the funded full projects.
Aim 1 is to attract, select and fund the most outstanding proposals with significant potential to benefit the SPORE in Skin Cancer and translational skin cancer research. Aim 2 is to support and integrate the selected pilot projects into the SPORE program with established processes to review and monitor progress. Women and underrepresented minorities will be strongly encouraged to participate. The DRP has a transparent peer-reviewed selection process that incorporates defined criteria for funding decisions. We require that DRP awardees be active in SPORE functions, use the SPORE Core resources, and provide formal written progress reports. Within the context of the DRP, we have established high standards and expectations of the investigators and have established a formal process to review and monitor progress of funded pilot projects. The DRP measures success by publications, extramural funding, stimulating new aims in existing SPORE projects, and elevation to full projects for the future.
Career Enhancement Program
Program Directors:
Jessie Villanueva, PhD
Lynn Schuchter, MD
The Career Enhancement Program (CEP) will recruit, train, and retain a diverse pool of early-career investigators who are committed to pursuing translational skin cancer research. The CEP will primarily assist junior faculty in their career development, but more established investigators interested in refocusing their research efforts on melanoma/skin cancer could also be considered. Women and underrepresented minorities are strongly encouraged to participate. CEP awardees will receive funding for up to two years.







