Targeting Lung Cancer Vulnerabilities
University of Texas Southwestern Medical Center
Principal Investigator(s):

John Minna, MD

John Heymach, MD, PhD
- Principal Investigators Contact Information
- Overview
- Project 1: Targeting Metabolic Vulnerabilities in Lung Cancer
- Project 2: Targeting immune vulnerabilities in Lung Cancer
- Project 3: Targeting Vulnerabilities in the Fibrotic Extracellular Matrix (ECM) of Lung Cancers
- Project 4: Therapeutic Targeting of Replication Stress Vulnerabilities in Small Cell Lung Cancer
- Administrative Core
- Molecular Pathology and Tissue Resource Core
- Data Science Core (Biostatistics/Bioinformatics/Database)
- Developmental Research Program
- Career Enhancement Program
- Institutional SPORE Website
Principal Investigators Contact Information
John Minna, MD
Director, Hamon Center for Therapeutic Oncology Research
Professor, Internal Medicine and Pharmacology
University of Texas Southwestern Medical Center
6000 Harry Hines Blvd
NB8.206B/MC8593
Dallas, TX 75390
(214) 648-4900
John Heymach, MD, PhD
Professor and Chair, Department of Thoracic/Head and Neck Medical Oncology
UT MD Anderson Cancer Center
1500 Holcombe Blvd., Unit 432, PO Box 301402
Houston, TX 77030
(713) 792-6363
Overview
Targeting Lung Cancer Vulnerabilities. The University of Texas SPORE in Lung Cancer represents a unique collaboration between the University of Texas Southwestern Medical Center (UTSW) and the University of Texas MD Anderson Cancer Center (MDACC), both of which have outstanding strengths in lung cancer translational and clinical research. The overarching goal of the SPORE is to develop new therapeutic paradigms based on recently identified “vulnerabilities” acquired during lung cancer pathogenesis, including a molecular understanding of lung cancers in individual patients, and using this information to “personalize” therapy for each lung cancer patient. Thus, our strategy is to identify lung cancer “therapeutic quartets” which include: 1. a specific vulnerability; 2. the mechanism of action thus defining therapeutic target(s) for the vulnerability; 3. a deliverable treatment for the target(s); and 4. tumor molecular biomarkers for the vulnerability predicting specific therapies for each patient. The UT Lung Cancer SPORE builds on a 20-year productive history, incorporating recent advances made by our SPORE investigators and the rest of the lung cancer translational research community in the molecular and mechanistic understanding of tumor autonomous and microenvironment changes, acquired vulnerabilities, and important immuno-oncology effects. These advances include novel approaches to identifying and molecularly classifying vulnerabilities in lung cancer metabolomic changes, cancers immunologically “inert” to PD1/PD-L1 checkpoint blockade, the lung cancer fibrotic stroma (microenvironment), and tumorigenesis-induced replication stress. Our contributions also include preclinical human and mouse model systems for testing the different vulnerabilities, as well as large legacy molecular and clinically annotated preclinical model and clinical specimen datasets.
The SPORE is composed of 4 projects, all of which have Human Endpoints:1. Targeting metabolic vulnerabilities in lung cancer; 2. Targeting vulnerabilities in immunologically-inert lung cancer; 3. Targeting vulnerabilities in the fibrotic extracellular matrix (ECM) of lung cancers; and 4. Therapeutic targeting of oncogene-induced replication stress for tumor cell killing and anti-tumor immunity in small cell lung cancer (SCLC) (which includes a clinical trial targeting replication stress combined with immune checkpoint inhibition. There are three cores:A. Administrative (including patient advocates); B. Molecular Pathology and Tissue Resources; and C. Data Sciences, as well as strong Developmental Research and Career Enhancement Programs (DRP, CEP). Our SPORE features leading lung cancer multi-disciplinary clinical and laboratory scientists, a cadre of experienced patient advocates, and an outstanding publication record. Moving forward, this SPORE will provide information on newly identified lung cancer acquired vulnerabilities, biomarkers for personalizing individual patient therapy, and important preclinical and information to facilitate clinical translation that has the possibility of changing the face of lung cancer therapy.
Project 1: Targeting Metabolic Vulnerabilities in Lung Cancer
Project Co-Leaders:
Ralph DeBerardinis, MD, PhD (basic)
Kate O’Donnell, PhD (basic)
John Minna, MD (clinical)
Human lung tumors are metabolically distinct from adjacent lung tissue. It is unknown whether these reprogrammed activities predict clinical outcomes or represent meaningful therapeutic liabilities. The major bottleneck in understanding the clinical relevance of cancer metabolism has been a lack of data about human tumor metabolism in vivo. For the first time, we have overcome this bottleneck and used intra-operative infusions with 13C-glucose to define metabolic phenotypes in human non-small cell lung cancers (NSCLC). We reported that one NSCLC subset displays prominent import of lactate while another produces lactate from glucose. Our observation that lactate uptake via monocarboxylate transport protein-1 (MCT1) correlates with rapid disease progression in lung adenocarcinoma (LUAD) is the first and so far only metabolic flux phenotype demonstrated to predict clinical outcomes in any human cancer. Here we will expand the scope of metabolic analysis in human NSCLC, performing 13C infusions in more than 100 patients, assessing hundreds of metabolites in each tumor, and following clinical histories to identify new activities correlating with outcomes. In Specific Aim 1, tumors infused with 13C will be analyzed by imaging, quantitative histopathology, RNA sequencing and whole exome sequencing to understand relationships between these features and cancer metabolism. We will focus on finding metabolic features that correlate with reduced progression-free survival, under the rationale that such activities are attractive therapeutic targets to test in preclinical models. We will establish patient-derived xenografts (PDXs) from these tumors to test the importance of predictive metabolic activities for tumor growth and metastasis. While our open-ended metabolomics approach is designed to uncover novel therapeutic targets, we will specifically test whether inhibiting MCT1 reduces tumor growth and metastasis in mice. In Specific Aim 2 we will follow up on our observation that lung squamous cell carcinomas (LSCCs) require lactate export for maximal growth. We will test whether genetic or pharmacological inhibition of novel molecular components of MCT4-mediated lactate export suppresses LSCC growth in mouse models and PDXs. Specific Aim 3 will examine metabolic crosstalk among cancer cells and several important immune cell populations in the tumor microenvironment (TME) in mice and humans. We will test the hypothesis that lactate metabolism impacts these metabolic exchanges and that blocking lactate transport enhances the efficacy of immune checkpoint blockade therapy. Overall, these efforts will produce the most detailed and clinically-relevant view of NSCLC metabolism to date. The ability to combine our ongoing study assessing metabolic flux in human NSCLC with large legacy clinical datasets ideally positions us to understand the relationship between tumor metabolism and cancer progression, and to advance high-priority therapeutic targets into clinical trials. The immediate Human Endpoint of this project is the direct, detailed examination of tumor metabolism and TME in lung cancer patients following 13C-glucose infusions, while later Human Endpoints will involve therapeutic interventions targeting MCT1 and MCT4. As another Human Endpoint we have added the conduct of an early phase clinical trial of Selinexor (XPO1 inhibitor, KPT-330) in KRAS mutant relapsed NSCLC in combination with docetaxel (NCT 03095612) and determination in patients and preclinical models of the impact of the new targeted therapy (XP01 inhibitor) on KRAS mutant lung cancer metabolism as a sub aim to Specific Aim 1.
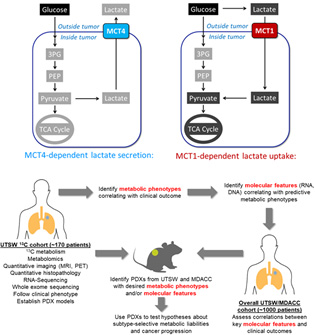
Top, metabolic phenotypes in NSCLC include MCT4-mediated lactate secretion and MCT1-mediated lactate uptake. Bottom, experimental and informatics workflow for Aim 1. Tumors infused with 13C will be analyzed by imaging, quantitative histopathology, RNA sequencing and whole exome sequencing to understand relationships between these features and cancer metabolism. We will focus on finding metabolic features that correlate with reduced progression-free survival, under the rationale that such activities are attractive therapeutic targets to test in preclinical models. We will establish patient-derived xenografts (PDXs) from these tumors to test the importance of predictive metabolic activities for tumor growth and metastasis.
Project 2: Targeting immune vulnerabilities in Lung Cancer
Project Co-Leaders:
John Heymach, MD, PhD (basic)
David Gerber, MD (clinical)
Ferdinandos Skoulidis, MD, PhD (Clinical)
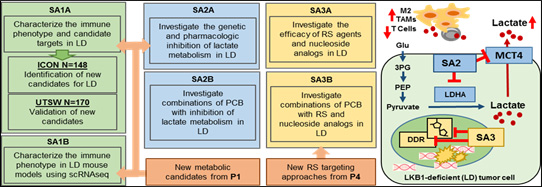
Immunotherapy with PD-1/PD-L1 checkpoint blockade (PCB) given alone or with chemotherapy now represents the standard first-line treatment for NSCLC patients with wild-type (wt) EGFR and ALK. This is a major advance, but the majority of NSCLC patients do not have an objective response to PCB. The molecular determinants of PCB resistance are incompletely understood, although low tumor mutation burden and PD-L1 levels predict some cases. Recently, we reported that LKB1 deficient (LD) tumors resulting from mutations or deletions in the STK11/LKB1 gene are associated with an inert or “cold” immune phenotype and represent the largest genomically-define subgroup with primary resistance to PCB, accounting for more than 30% of PCB resistance in lung adenocarcinoma. The LKB1 protein is a master regulator of metabolism, energetic balance, and nucleotide stores. Recent publications from our group and others indicate that LD tumors have a distinct metabolic phenotype that includes enhanced lactate production, vulnerability to targeting intracellular nucleotide pathways, and increased replicative stress (RS). These features may contribute to the “cold” immune phenotype. The primary goals of Project 2 (P2) are to investigate new therapeutic approaches for targeting LD NSCLC and enhancing antitumor immunity, with a focus on targeting the lactate pathway and RS. This focus integrates the immunotherapy focus of Project 2 with studies in the other SPORE Projects and provides multiple Human Endpoints for the SPORE Projects. We hypothesize that: 1) Enhanced lactate production or secretion contributes to the “cold” immune phenotype in LD NSCLC; 2) LD NSCLC will be preferentially vulnerable to targeting RS; and 3) targeting RS and/or lactate pathways may enhance antitumor immunity and response to PCB. To test these hypotheses,
Specific Aims:
Aim 1: We will comprehensively characterize the immune phenotypes of LD NSCLC using two sets of resected tumors: the MD Anderson ICON cohort and a validation cohort from UTSW which have undergone metabolic labeling in P1. We will also examine immune cells in greater detail using single cell RNA sequencing.
Aim 2: We will test whether targeting the lactate pathway using inhibitors of MCT4 and LDHA can reverse LD-associated immunosuppression and enhance PCB efficacy.
Aim 3: We will target RS using inhibitors of ATM, ATR, and the nucleoside analog 6-thio-dG in collaboration with P4, alone or in combination with PCB. Significance: LD NSCLC tumors have a “cold” immune phenotype and frequent primary resistance to PCB or PCB/chemotherapy. This patient population is larger than EGFR-mutant NSCLC and comparably sized to metastatic pancreatic cancer. P2 aims to leverage our unique set of clinical and preclinical resources to develop more effective therapeutic approaches for LD NSCLC patients, which can then translated by our group and others into the clinic. P2 also provides the opportunity to spearhead a new paradigm of genomically-guided, tailored immunotherapy for PCB-resistant tumors.
Project 3: Targeting Vulnerabilities in the Fibrotic Extracellular Matrix (ECM) of Lung Cancers
Project Co-Leaders:
Don Gibbons, MD, PhD (Clinical/Applied Leader)
Jonathan Kurie, MD (Basic Co-Leader)
Substantial therapeutic advances have been made in the treatment of non-small cell lung cancer (NSCLC) based on the incorporation of immune checkpoint inhibitors of the PD-1/PD-L1 axis. Unfortunately, the majority of patients do not benefit from this single-agent approach, either due to primary or acquired resistance mechanisms. There is a knowledge gap about the interplay between the tumor-infiltrating immune cells and the dynamic changes in the extracellular matrix and tumor microenvironment that may drive an immunosuppressive microenvironment, which translates into a major unmet therapeutic need. The members of our multidisciplinary team (Gibbons, Kurie, Cascone, Yamauchi, Dalby, Wistuba, and Wang) have a track record of productivity in studying tumor matrix dynamics and immune factors in the microenvironment during lung cancer progression and in response to immune therapy. The investigators bring expertise in mouse modeling of human lung cancer, clinical oncology, immunotherapy, molecular pathology of lung cancer, drug development and bioinformatics/biostatistics. We have developed preliminary data from analysis of human lung cancer specimens and preclinical genetically engineered mouse models (GEMMs) of NSCLC that the deposition and crosslinking of the collagen matrix suppresses intratumoral surveillance by immune cells. Based on these findings, we hypothesize that extracellular matrix deposition and collagen crosslinking in the tumor microenvironment suppress tumor-infiltrating immune cells, facilitating tumor invasion and metastasis. Through parallel study of pre-clinical models and assessment of patient samples, we will address this hypothesis and explore clinical translational opportunities by:
Specific Aims:
Aim 1: evaluating the role of collagen crosslinking enzymes and a fibrotic extracellular matrix on the immune cell profile of murine and human lung tumors;
Aim 2: determining how treatment with immune checkpoint inhibitors (ICI, e.g. anti-PD-(L)1 and/or anti-CTLA-4) alters extracellular matrix composition and crosslinking in murine and patient tumors; and
Aim 3: testing the efficacy of agents that block fibrotic collagen crosslinking or collagen-mediated T cell suppression, with or without immune checkpoint inhibitors to reverse the suppression of a productive intratumoral immune response in preclinical models. The Human Endpoint is demonstrated in our analysis of key ECM features in: A. SPORE tumor samples; B. 150 paired surgical tumor/normal samples and >500 blood samples from previously untreated patients with stage IB-IIIA NSCLC from the MDACC ImmunogenomiC prOfiling of NSCLC (ICON) study; C. 44 paired tumor/normal samples from patients treated on the MDACC NEOSTAR study of neoadjuvant ICI for resectable stage I-IIIA NSCLC; and D. tumors from patients after treatment with the multi-kinase inhibitor nintedanib.
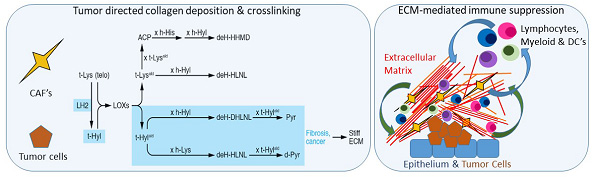
Left shows tumor cell and cancer associated fibroblast pathways that produce collagen crosslinking in the extracellular matrix (ECM) of the tumor microenvironment, with subsequent effects on the tissue properties. Right shows the bi-directional interactions between epithelial cells (normal or cancerous) and ECM, and ECM with the immune compartment. These interactions can produce suppression of effective immune surveillance.
Project 4: Therapeutic Targeting of Replication Stress Vulnerabilities in Small Cell Lung Cancer
Project Co-Leaders:
Lauren Byers, MD (Clinical/Applied Leader)
Jerry Shay, PhD (Basic Co-Leader)
The goal of this project is to develop novel, synthetic lethal, replication stress related, approaches for SCLC patients based on their tumor’s molecular profiles. There are two major unmet needs for SCLC therapy: first, the majority of patients will experience primary or acquired resistance to clinically available chemotherapy and immunotherapy; and second, despite new data supporting molecularly and biologically distinct subsets of SCLC, there are no established approaches for biomarker-driven personalized treatments (such as is standard for targetable oncogenic drivers in NSCLC). Many of the currently “undruggable” oncogenic changes that are common in SCLC promote tumorigenesis through replication stress (RS) and genomic instability. These “hallmarks of cancer” lead to high tumor mutational burden and chromosomal aberrations. SCLCs have a continuous high degree of RS due to the universal loss of p53 and RB1 function, and frequent activation of oncogenes such as MYC. In the presence of RS, cancer cells depend on RS responses (RSR), which are a branch of DNA damage repair responses (DDR), to resolve DNA damage and stalled replication forks. Several RSR inhibitors are in clinical development, and we have preliminary findings in preclinical models indicating that SCLC lines, xenografts, and genetically engineered mouse models (GEMMs) are sensitive to agents that target RSR or that increase RS or genomic instability beyond tolerable levels. The latter group includes aurora kinase inhibitors (which induce mitotic catastrophe in the setting of high RS levels, as well as directly inhibiting DNA repair and decreasing replication fork stability) and 6-thio-dG (distinct from the well-known drug 6 thioguanine) which is incorporated by telomerase positive cells, leading to telomere disruption, and increased RS. Furthermore, we have recently found that therapeutic targeting of RS (e.g., by inhibitors of Chk1 or PARP) increases cytoplasmic DNA, resulting in activation of the innate immune response (via the Stimulator of Interferon Genes [STING] pathway), immune-mediated cytotoxicity, and enhanced response to immune checkpoint blockade. We hypothesize that: 1. RS targeting (through Aurora Kinase inhibition or telomere disruption) will lead to synthetic lethality in molecularly-defined subsets of SCLC (Aim 1); and 2. that RS targeting will enhance response to immune checkpoint blockade in immunotherapy-refractory subsets of SCLC via activation of the innate immune system (Aims 2 and 3). Specifically, we expect that RS-targeted therapies will lead to replication catastrophe and cell death in oncogene-driven lung cancers and enhance responses to immune checkpoint blockade (e.g., anti-PD-L1). We will test our hypotheses in an extensive panel of molecularly characterized SPORE lung cancer cell lines, patient derived xenografts (PDXs, tumor and circulating tumor cell (CTC)-derived), GEM-derived animal models (Aims 1 and 2), and in a Human Endpoint using clinical specimens from an investigator-initiated Phase 2 clinical trial (Aim 3) of Aurora Kinase B inhibition with AZD2811 in relapsed extensive-stage SCLC.
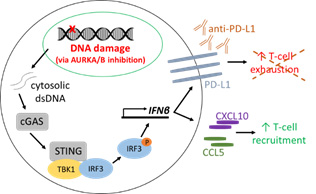
A. Replication stress targeting via Aurora Kinase inhibitors activates the STING pathway, leading to increased T-cell recruitment and PD-L1 levels.
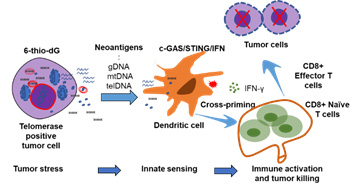
B: Telomerase-mediated replication stress-targeted therapy on immune modulation function
Administrative Core
Core Directors:
John D. Minna, MD (Core Director)
John V. Heymach, MD, PhD (Core Co-Director)
The Lung Cancer SPORE Administrative Core provides scientific and administrative oversight for all of the SPORE Projects and Cores and the Developmental Research (DRP) and Career Enhancement (CEP) Programs. It also provides the main interface with patient advocates, NCI program staff, external and internal advisory boards, other Lung Cancer SPOREs, and other collaborating institutions. The SPORE Administrative Core is co-Directed by the SPORE MPIs Drs. John D. Minna and David E. Gerber at UTSW, and Drs. Jack A. Roth, and John V. Heymach at MDACC. The MPI plan details how they will work together and which aspects of the SPORE administration they will focus on. As part of a planned transition, Drs. Gerber and Heymach will assume full responsibility for the Administrative Core in years 3-5.
Specific Aims:
Aim 1: Direct the overall scientific quality and administrative management of the SPORE and ensure effective communication between SPORE investigators and Patient Advocates; facilitate integration of Research Projects and Cores within and between UTSW and MDACC; encourage and facilitate multidisciplinary collaborations and cooperation (including Patient Advocates), as well as data and resource sharing.
Aim 2: Prepare budgets, monitor budget expenditures, and implement corrective actions to stay within budget.
Aim 3: Coordinate all SPORE investigator meetings, including monthly SPORE works in progress (WIPs) videoconference/WebEx meetings, and Internal and External Advisory board meetings.
Aim 4: Coordinate access to SPORE Core resources and oversee data quality control and validation determined by review of the Data Sciences Core.
Aim 5: Ensure implementation of all federal, state, and institutional regulatory compliances.
Aim 6: Oversee SPORE DRP and CEP activities by advertising DRP and CEP grant opportunities, and conducting DRP and CEP grant review processes to ensure identification of the projects with the combination of the best science, innovation, and potential for clinical translation; work with DRP and CEP applicants to help them use SPORE resources and refine their projects for optimum translational potential, which will include identification of qualified mentors for DRP and CEP awardees; establish and carry out policies for recruitment of women and minorities to be involved scientifically in the SPORE and DRP and CEPs.
Aim 7: Enhance recruitment of women and minority patients to participate in SPORE-related clinical trials.
Aim 8: Encourage and facilitate horizontal and vertical collaboration of SPORE discoveries and investigators both within UTSW and MDACC and with other institutions.
Aim 9: Coordinate and maintain institutional commitments to the SPORE at UTSW and MDACC.
Aim 10: Communicate and consult with the NCI program director frequently to ensure adherence to all reporting requirements, including high-quality progress reports.
Aim 11: Coordinate and facilitate SPORE interactions with pharmaceutical and biotech industry partners.
Aim 12: Coordinate and facilitate Patient Advocate involvement in the SPORE.
Aim 13: Coordinate and facilitate resolution of scientific disputes.
Molecular Pathology and Tissue Resource Core
Core Directors:
Ignacio Wistuba, MD
Justin Bishop, MD
The Molecular Pathology and Tissue Resources Core will provide routine and innovative tissue resources and materials essential for achieving the aims of the SPORE projects. Routine materials include tumors, non-malignant lung specimens, tumor cell lines, and clinical annotation, all of which have been obtained with informed consent and measures to ensure patient confidentiality. Over 6,000 tumors including tumors on clinically annotated tissue microarrays (TMAs), 300 lung cancer cell lines of all histologic types, and over 180 lung tumor patient-derived and circulating tumor cell-derived xenografts (PDXs/CDXs) are available to investigators. Many of these samples have been distributed to our SPORE investigators, as well as other investigators at our own and outside institutions (including other SPOREs). These partnerships have fueled multiple lung cancer translational research collaborations, and have generated over 380 publications in the last ten years. In addition to important clinical and histologic annotation, the genomic, molecular, and immune microenvironment profiling of these samples provide important Human Endpoints for correlative data and synergy, both within the UT Lung SPORE and as components of other lung cancer translational research. Given the increasing importance of immunotherapy for lung cancer, the Pathology Core has developed methods to provide state of the art immune profiling annotation. Additionally, the Core has participated in collection of patient samples from the MDACC ImmunogenomiC prOfiling of NSCLC (ICON) and NEOadjuvant STudy of induction checkpoint blockade for Resectable stage I-IIIA NSCLC (NEOSTAR) clinical trials. Our Aim 1 is to collect, process, store, catalog and distribute tissues, cell lines and blood specimens, both malignant and non- malignant, tumor xenografts, and relevant clinico-pathologic and molecular data, as requested by the various component projects of the SPORE program; we will use IRB-approved protocols, informed consent, and measures to protect subject confidentiality. Aim 2 is to develop and utilize innovative and routine tissue and cell line resources that will aid in the successful completion of the SPORE program aims; these include development of new tumor cell lines, additional lung cancer TMA resources, molecular analysis of tumors and liquid biopsy, and comprehensive immune profiling of tissues. Aim 3 is to perform, interpret, analyze, and deposit tissue-based molecular and immune analysis methodologies in close collaboration with the component Projects, DRP, CEP, and Data Sciences Core of the SPORE program to achieve their approved aims. Within these aims, the Pathology Core will play a crucial role in promoting vertical and horizontal collaborations among our own SPORE investigators, investigators at other Lung Cancer SPORE sites, and other investigators at our own and other institutions. All of our SPORE projects will use Core B materials and services. Heavy utilization of our routine and innovative materials, as well as close interactions with the SPORE investigators, will greatly aid the successful completion of the aims of our SPORE proposal.
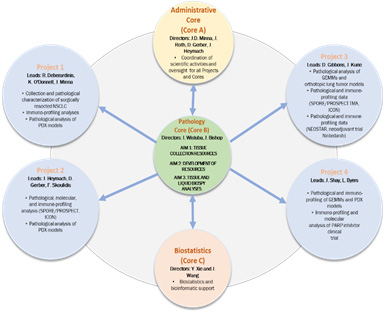
Data Science Core (Biostatistics/Bioinformatics/Database)
Core Directors:
Yang Xie, PhD
Jing Wang, PhD
The research proposed by our SPORE in Lung Cancer encompasses a broad range of activities, including studies in clinically annotated patient tumor samples, tumor cell lines, xenografts, mouse models, and clinical trials. These studies generate many different types of data, including clinical, histologic, genome wide molecular (mutation, expression), proteomic, biochemical, immunohistochemical, drug and immune response phenotype, metabolomic, and tumor environmental. The Data Sciences Core provides comprehensive expertise to ensure the statistical integrity, data integrity, data sharing capability, and data analysis accuracy of the studies performed by the SPORE, which are conducted at UTSW and MDACC. The Core has a Director at each institution (Y. Xie, UTSW, and J. Wang, MDACC) and the flexibility to match personnel to the evolving needs of existing SPORE Projects, and Developmental Research and Career Enhancement Program (DRP, CEP) Projects. Members of the Core participate in both monthly all-SPORE Project and Core meetings, and in the specific Data Sciences SPORE video/WebEx conferences linking researchers at UTSW in Dallas and MDACC in Houston, TX, ensuring that proper consideration is taken of biostatistics and data management issues during all SPORE work. The Data Sciences Core will develop and maintain systems for data storage, retrieval, analysis, and sharing. It will provide an interface for all SPORE investigators to easily and freely exchange data and information. The Core will provide analyses to allow others outside the SPORE to have appropriate access to SPORE datasets, and to be able easily to independently reproduce and validate biostatistical and computational analyses. The Core services include innovative, unique, and sometimes customized approaches to solving the data analysis and interpretation challenges of the modern data-centric research laboratory. The Core Specific Aims are: Aim 1: To provide valid statistical designs for laboratory research, clinical trials and translational experiments arising for SPORE research; Aim 2: To oversee and conduct the innovative statistical modeling, simulations, data analyses and data integration needed by the Projects, DRP and CEP, and Pathology Core, to achieve their specific aims; Aim 3: To ensure that all complex molecular, biologic, and clinical datasets are protected for confidentiality, analyzed, shared among SPORE investigators and collaborators, and appropriately deposited into publically accessible databases as required, using valid and innovative bioinformatics methods; Aim 4: To develop and maintain a secure, web-accessible site for SPORE research data integration and storage linked to an extensive tissue repository of clinically and molecularly annotated archived patient samples, tumor grafts, tumor and normal cell lines, and relevant mouse models of lung cancer. To develop and maintain centralized deposits from the literature of lung cancer-relevant datasets in a web site (“Lung Cancer Explorer”) to support SPORE investigators and, more broadly, the research community. To provide data-related analyses and documents for publication (such as “Sweave”) that allow the research community to independently reproduce and validate our analyses.
Developmental Research Program
Program Directors:
John D. Minna, MD
John Heymach, MD, PhD
As mandated by NCI SPORE Guidelines, the University of Texas Special Program of Research Excellence (SPORE) has a Developmental Research Program (DRP). The SPORE DRP is funded by a combination of NCI SPORE grant support ($50,000 direct costs), and institutional support from UTSW and UTMDACC ($150,000 direct costs) for a total budget of $200,000. With average awards of $25,000, this funding permits ~8 DRP Awards per year.
Specific Aims:
Aim 1: Provide Funding for Developmental Research Projects;
Aim 2: Use expertise of members of the SPORE External and Internal Scientific Advisory Boards, and Senior Executive Committee to identify high-impact projects with potential lung cancer clinical translational utility;
Aim 3: Build on the existing well-structured SPORE mentorship platform for Development Research Projects;
Aim 4: Build on the existing framework for direct communication between basic and clinical scientists, within and outside UTSW and UTMADCC, across disciplines, and training and guidance of a new generation of translational lung cancer researchers;
Aim 5: Facilitate development and transition of these successful projects into competitive applications for extramural peer-reviewed funding;
Aim 6: Build on the existing well-structured SPORE DRP to ensure that scientific advances are translated into the clinic to benefit patients with lung cancer.
The overall DRP goals are to fund UTSW, MDACC personnel, and extra-institutional candidates with MD or PhD degrees at all academic levels who want to pursue a career in lung cancer research and who have prepared an application with high potential for significant impact on lung cancer pathogenesis, diagnosis, prevention, prognosis, or therapy. As part of this effort, awardees will have access to Lung Cancer SPORE Core resources (such as preclinical models, Molecular Pathology, Data Sciences), and access to mentorship by experienced lung cancer investigators and patient advocates. DRP Awardees will bring new and innovative ideas and approaches to lung cancer translational research and be trained in identifying major lung cancer problems to attack, as well as new approaches, concepts, technologies, data gathering and analyses for understanding lung cancer biology, pathogenesis, diagnosis and therapy. The DRP is run by the SPORE MPIs from UTSW and MDACC (Minna, Gerber, Roth, Heymach). Applications are scored by the SPORE External Advisory Board (EAB) based on the applicant’s ability, significance of the project, innovation, approach, research Environment, and potential of project. Special efforts are made to recruit women and minority applicants. Over the 20-year history of this SPORE, the DRP has been highly successful, with a large number of awards leading to important publications, providing preliminary data for other grants, assisting with faculty promotions, and progression of DRP recipients to major roles in SPORE Projects and Cores.
Career Enhancement Program
Program Directors:
John D. Minna, MD
John Heymach, MD, PhD
As mandated by NCI SPORE Guidelines, the University of Texas Special Program of Research Excellence (SPORE) has a Career Enhancement Program (CEP). The SPORE CEP is funded by a combination of NCI SPORE grant support ($50,000 direct costs) and Institutional support from UTSW and MDACC ($150,000 direct costs) for a total budget of $200,000. With average awards of $25,000, this permits ~8 CEP Awards per year.
Specific Aims:
Aim 1: Maintain lung cancer translational research excellence at UTSW and MDACC by recruitment of highly innovative and talented clinical, applied, and basic scientists at all levels.
Aim 2: Promote the career enhancement of highly qualified clinical oncologists, applied, and basic scientists, both within UTSW and MDACC and outside of these institutions, with the ability to rapidly translate their findings into clinically applicable utility for lung cancer.
Aim 3: Attract candidates with prior experience in cancer at other disease sites who want to acquire expertise in lung cancer translational research.
Aim 4: Promote transition of CEP awardees from mentored investigators to successful independent lung cancer translational research scientists including helping them prepare other peer-reviewed funding applications.
The overall CEP goals are to fund UTSW, MDACC young faculty with MD or PhD degrees (senior post- doctoral fellow transitioning to faculty, Instructor, Assistant and Associate Professor level) who want to pursue a career in lung cancer research and who have prepared an application with high potential for significant impact on lung cancer pathogenesis, diagnosis, prevention, prognosis, or therapy. As part of this program, awardees will have access to Lung Cancer SPORE Core resources (such as preclinical models, Molecular Pathology, Data Sciences), and access to mentorship by experienced lung cancer investigators and patient advocates. CEP Awardees will bring new and innovative ideas and approaches to lung cancer translational research; in turn, they will receive training in new approaches, concepts, technologies, data gathering and analyses for understanding lung cancer biology, pathogenesis, diagnosis and therapy. The CEP is run by the SPORE MPIs from UTSW and MDACC (Minna, Gerber, Roth, Heymach). Applications are scored by the SPORE External Advisory Board (EAB) based on the applicant’s ability and the project’s significance, innovation, approach, research environment, and potential impact. Special efforts are made to recruit women and minority applicants. Over the 20-year history of this SPORE, the CEP has been very successful, leading to a large number of awardees whose careers have been enhanced by the awards, including publications, generation of preliminary information for obtaining other grants, academic promotion, and progression to playing major roles in SPORE Projects and Cores.
Institutional SPORE website
https://www.utsouthwestern.edu/labs/minna/collaborations/developing-personalized-medicine.html







