SPORE in Breast Cancer
University of North Carolina
Principal Investigator(s):

Charles Perou, PhD

Lisa Carey, MD
- Principal Investigator(s) Contact Information
- Overview
- Project 1: Carolina Breast Cancer Study: Linking Tumor Biology to Social Determinants
- Project 2: Determinants of Response and Resistance to DNA-Damaging Radiation Plus Immunotherapy Combinations in Triple Negative Breast Cancer
- Project 3: Development of Novel Therapies and Biomarkers for Triple Negative Breast Cancer Patients
- Project 4: Activity of CAR-T Cell Therapy for Patients with Metastatic Triple Negative Breast Cancer
- Core A: Administrative
- Core B: Biostatistics & Bioinformatics Core
- Core C: Pathology and Immunobiology
- Developmental Research Program
- Career Enhancement Program
- Institutional website
Principal Investigator(s) Contact Information
Charles Perou, PhD
The May Goldman Shaw Distinguished Professor of Molecular Oncology
Department of Genetics, and Pathology & Laboratory Medicine; Co-Leader, UNC Lineberger Breast Cancer Research Program
UNC Lineberger Comprehensive Cancer Center
Marsico Hall 5111, 125 Mason Farm Road, CB #7599, Chapel Hill, North Carolina, 27599
(919) 843-5740
Lisa Carey, MD, ScM
L. Richardson and Marilyn Jacobs Preyer Distinguished Professor for Breast Cancer Research; Deputy Director of Clinical Sciences, UNC Lineberger
UNC Lineberger Comprehensive Cancer Center, General Administration Suite 10-000, 450 West Drive, CB#7295, Chapel Hill, North Carolina, 27599
(919) 843-6814
Overview
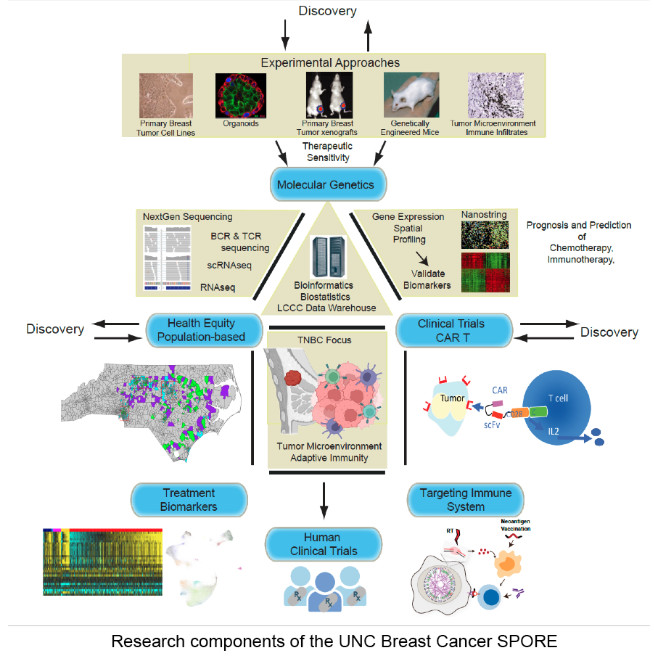
The UNC Breast Cancer SPORE conducts research that translates laboratory discoveries into new and more effective breast cancer treatments, with the goal to improve the outcome for breast cancer patients in North Carolina and across the nation. UNC Lineberger Comprehensive Cancer Center’s strengths in bioinformatics, immunogenomics, proteomics and population-based cohort studies supports the UNC Breast Cancer SPORE’s clinical, public health and basic researcher teams. The SPORE researchers’ work includes the continuation of two studies supported by previous SPORE funding and the launch of two new studies. Specifically, the research projects will focus on racial and age-related disparities in outcomes, biomarkers, immunotherapy and triple negative breast cancer that is resistant to conventional treatments:
- Project 1: Carolina Breast Cancer Study: Linking Tumor Biology to Social Determinants
- Project 2: Determinants of Response and Resistance to DNA-Damaging Radiation Plus Immunotherapy Combinations in Triple Negative Breast Cancer
- Project 3: Development of Novel Therapies and Biomarkers for Triple Negative Breast Cancer Patients
- Project 4: Activity of CAR-T Cell Therapy for Patients with Metastatic Triple Negative Breast Cancer
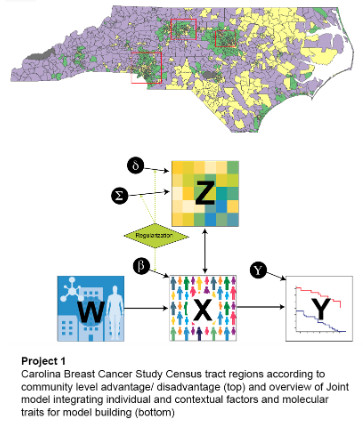
Project 1: Carolina Breast Cancer Study: Linking Tumor Biology to Social Determinants
Project Co-Leaders:
Melissa Troester PhD, MPH (Basic Science)
Katherine Reeder-Hayes MD, MBA, MSCR (Clinical)
Black women experience worse breast cancer outcomes, with tumor biology, individual exposures, community-level, and structural determinants all contributing to this disparity. In the Carolina Breast Cancer Study phase 4, we are evaluating tumor biology in context of a multi-level cells-to-society model to identify predictors of recurrence and survival and to identify new interventions allowing precision medicine in breast cancer. Specifically, our focus is on developing better understanding of multilevel determinants of breast cancer outcome disparities and interventions to increase health equity in the context of precision medicine.
Aim 1: Identify biological tumor features (proliferation, p53 status, DNA repair, immune response, and stromal reactivity) that vary according to individual- and community-level factors under a cells-to-society framework for health equity.
Aim 2: Identify relationships between tumor biology, access to care, and community-level variables (exposures) with breast cancer recurrence/survival (outcomes) in a diverse study population (Carolina Breast Cancer Study 3 & 4).
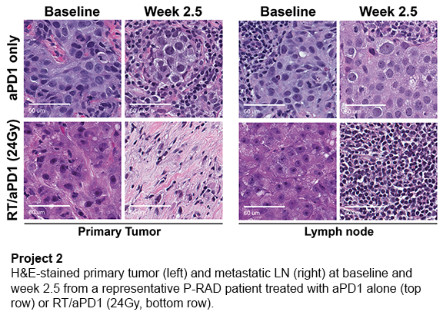
Project 2: Determinants of Response and Resistance to DNA-Damaging Radiation Plus Immunotherapy Combinations in Triple Negative Breast Cancer
Project Co-Leaders:
Gaorav Gupta, MD, PhD (Clinical)
Benjamin Vincent, MD (Basic Science)
Combining immune checkpoint inhibition with chemotherapy has transformed outcomes for patients with early stage triple negative breast cancer (TNBC). However, treatment resistance and treatment-related toxicities persist as dual challenges that must be overcome with new strategies to guide personalized chemo-immunotherapy combinations. We are investigating the role of DNA damage-induced necroptosis (inflammatory cell death) as a determinant of response to radiation therapy plus anti-PD1 (aPD1) immune checkpoint inhibitor combination therapy. We use TNBC genetically engineered mouse models and correlative biomarker analyses of pre/post-treatment primary tumor, metastatic lymph node, and blood specimens from an ongoing clinical trial of preoperative aPD1 therapy, with or without radiation therapy, in early-stage TNBC with lymph node metastasis.
Aim 1: Assess necroptosis as a predictive biomarker of response to radiation therapy/aPD1 combination therapy in triple negative breast cancer.
Aim 2: Evaluate therapeutic neoantigen vaccination as a strategy to overcome ion therapy/aPD1 resistance induced by necroptosis deficiency.
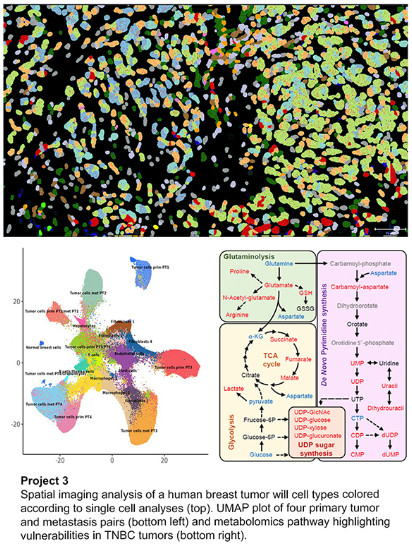
Project 3: Development of Novel Therapies and Biomarkers for Triple Negative Breast Cancer Patients
Project Co-Leaders:
Charles M. Perou, PhD (Basic Science)
Lisa A. Carey, MD, ScM (Clinical)
Triple negative breast cancer (TNBC) is a heterogenous clinical group that shows significant variation based upon histology, gene expression patterns, response to therapy, and even for patient survival outcomes. We are developing and testing new combination therapies in the lab that look promising for TNBC, and that will be guided by biomarker-based patient selection. Specifically, we evaluate drug treatments including immunotherapy such as anti-PD1 and CD40LG agonist antibody to boost B cell and/or dendritic cell function, and combinations containing drugs that inhibit macrophage function. Lastly, we have identified a unique metabolic vulnerability in TNBC, namely dependence upon pyrimidine biosynthesis, that may be a future therapeutic target.
Aim 1: Evaluation of novel immune therapy combinations for triple negative breast cancer.
Aim 2: Targeting triple negative breast cancer metabolism as a therapeutic strategy.
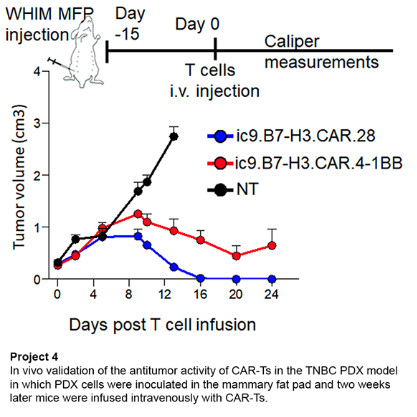
Project 4: Activity of CAR-T Cell Therapy for Patients with Metastatic Triple Negative Breast Cancer
Project Co-Leaders:
E. Claire Dees, MD, ScM (Clinical)
Gianpietro Dotti, PhD (Basic Science)
Jonathan Serody, MD (Translational Science)
Triple negative breast cancer (TNBC) remains a tumor with poor prognosis even in the era of immune checkpoint inhibitors. Our focus is on chimeric antigen receptor T cells (CAR-Ts) to target TNBC. We are currently targeting the B7-H3 antigen that is abnormally expressed in several solid tumors, including TNBC, but has limited expression in normal tissues. We are conducting a Phase I clinical trial to test the safety and antitumor activity of a novel CAR-T therapy in patients with TNBC. We are also studying new approaches to generating even more effective approaches to T cell-based immunotherapy.
Aim 1: Conduct a Phase I clinical study testing safety and effectiveness of autologous B7-H3.CAR-Ts in patients with TNBC.
Aim 2: Develop preclinical models of TNBC to test dual CAR-Ts targeting B7-H3 and CPSG4 and to engineer dual CAR-Ts to express the CCR2b chemokine receptor, which promotes CAR-T cell trafficking through the blood-brain-barrier when brain metastases produce the CCL2 chemokine.
Aim 3: Assess the mechanism and antitumor activity of Th/Tc17 CAR-Ts activated by cGAMP in modulating the breast cancer tumor microenvironment and enhancing the expansion of CAR-Ts in the tumor microenvironment.
Core A: Administrative
Core Directors:
H. Shelton Earp III, MD
Lisa A. Carey, MD, ScM
Charles M. Perou, PhD
The Administrative Core supports an exceptional team of breast cancer translational researchers who are exploring approaches to convert basic science discoveries into new and more effective breast cancer treatments. The Administrative Core provides scientific, administrative and financial oversight of all components of the UNC Breast Cancer SPORE. This includes coordinating input from leadership, advocates, and the External Advisory Board, with the objective of monitoring, evaluating, and supporting SPORE major and pilot projects, recruitment, career enhancement, and mentoring, and facilitating interactions with the NCI SPORE program and other NCI SPOREs.
Core B: Biostatistics & Bioinformatics Core
Core Directors:
Naim Rashid, PhD
Katherine Hoadley, PhD
The Biostatistics & Bioinformatics Core, led by faculty geneticists, statisticians, and biostatisticians, provides support for statistical and bioinformatics analysis of complex data sets and utilizes existing shared resources supported by the UNC Lineberger Comprehensive Cancer Center. It is an integral partner with each project. Complementary skills possessed by the Biostatistics & Bioinformatics Core faculty support the application and development of approaches for to cross-platform data integration and high-dimensional biomarker discovery to optimize clinical decision-making.
Core C: Pathology and Immunobiology
Core Directors:
Benjamin Calhoun, MD, PhD
Yisong Wan, PhD
The Pathology and Immunobiology Core supports all projects by providing a coordinated, quality-controlled, quality-assured facility for the procurement, processing, storage, and distribution of human biologic specimens. Specifically, it provides centralized tissue and specimen banking, receipt and processing of breast-related human specimens, including: freshly procured, snap-frozen and formalin-fixed paraffin embedded block specimens, preparation of tissue microarrays, cell microarrays, morphological evaluation and morphology-based assays, assay development and training, immunogenetics, flow-cytometry, and digital imaging and image analysis for spatial quantification of molecular analytes in intact specimens.
Developmental Research Program
Program Director:
H. Shelton Earp III, MD
The Developmental Research Program has promoted novel breast cancer research across the population, translational and clinical arenas. This program includes mechanisms for stimulating grant applications, rigorously evaluating proposals, selecting projects with advocate input, and monitoring progress. Evaluation and selection processes include a well-established, independent peer review that includes patient advocates and UNC Lineberger Community Outreach and Engagement Community Advisors.
Career Enhancement Program
Program Director:
Lisa A. Carey, MD, ScM
We are focused on identifying and recruiting junior faculty who have demonstrated excellence in a discipline that can be applied productively to breast cancer translational research; and promoting career development by providing adequate start-up, access to translational infrastructure within the SPORE and UNC Lineberger, and diligent mentorship, guidance and support.







