UCLA SPORE in Brain Cancer
University of California, Los Angeles
Principal Investigator:

Linda Liau, MD, PhD, MBA
- Principal Investigator Contact Information
- Overview
- Project 1: Targeting immunotherapy-induced resistance with DC vaccination and immune modulation
- Project 2: Overcoming drug-induced resistance to intrinsic apoptosis in glioblastoma
- Project 3: Strategies against radiation-induced cellular plasticity in glioblastoma
- Administrative Core
- Biospecimen and Pathology Core (BiPC)
- Neuro-Imaging Core (NIC)
- Biostatistics and Bioinformatics Core (BBC)
- Developmental Research Program
- Career Enhancement Program
- Institutional SPORE Website
Principal Investigator Contact Information
Linda Liau MD, PhD, MBA
Professor and W. Eugene Stern Chair
Department of Neurosurgery
University of California, Los Angeles
300 Stein Plaza, Suite 564
Los Angeles, California 90095-6901
Tel: (310) 267-9449
Overview
The objectives of the UCLA SPORE in Brain Cancer are to contribute significantly to progress in the diagnosis, prognosis, and treatment of brain cancer with a particular focus on developing novel strategies to overcome the problem of treatment-induced resistance. In order to achieve these translational research goals of our program, we propose three main projects involving:
Project 1: Targeting immunotherapy-induced resistance with DC vaccination and immune modulation
Project 2: Overcoming drug-induced resistance to intrinsic apoptosis in glioblastoma
Project 3: Strategies against radiation-induced cellular plasticity in glioblastoma
These translational research projects will be supported by shared resource cores in administration, biospecimen/pathology, neuroimaging, and biostatistics/bioinformatics. Our program will also be responsive to SPORE themes by incorporating Developmental Research and Career Enhancement Programs in order to foster new approaches for assessing and treating brain cancer.
Project 1: Targeting immunotherapy-induced resistance with DC vaccination and immune modulation
Project Co-Leaders:
Robert M. Prins, PhD (Basic Science Leader)
Linda M. Liau, MD, PhD, MBA (Clinical Science Leader)
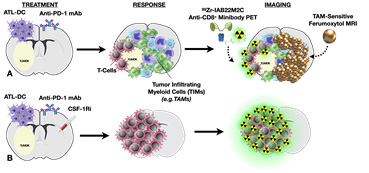
The overall goals of this project are to investigate mechanisms of immune evasion following treatment with dendritic cell (DC) vaccines, and to develop rational combinations of immunotherapeutic strategies to overcome the immunosuppressive milieu of the brain tumor microenvironment. We previously found that, in addition to inducing T-cell infiltration into brain tumors, DC vaccination + anti-PD1 blockade may also create a pro-inflammatory environment within the tumor that induces the immigration of immunosuppressive myeloid cells (TIM). TIM are phenotypically similar to the myeloid cells that attenuate the T-cell response to chronic viral infections, and may counteract the anti-tumor T-cell responses induced by DC vaccination. Therapies that target myeloid cells within the tumor microenvironment represent a promising new strategy. As such, inhibition of these myeloid cells using a CSF-1R inhibitor, in conjunction with autologous tumor lysate-pulsed DC vaccination (ATL-DC) and PD-1 mAb blockade, resulted in significantly prolonged survival in tumor-bearing animals with large, well-established intracranial gliomas. Our hypothesis is that myeloid cells mediate adaptive immune resistance in response to T-cell activation induced by immunotherapy. We have planned a series of novel pre-clinical studies to re-polarize myeloid cells, to optimize how the timing and sequence of immunotherapy can influence ant-tumor immunity, and a new clinical trial to test the first-in-human combination of a new brain penetrant CSF-1R inhibitor (CSF-1Ri; PLX3397, Daiichi-Sankyo) with DC vaccination and PD-1 mAb blockade (Pembrolizumab, Merck) in patients with newly diagnosed GBM. A better understanding of the biology of these cellular interactions will provide insight into more effective ways to induce therapeutic anti-tumor immune responses for this deadly type of brain tumor.
Aim 1: To identify the optimal timing and sequence by which immunotherapy alters the local tumor-infiltrating immune response in syngeneic murine glioma models and in recurrent glioblastoma patients.
Aim 2: To conduct a new first-in-human Phase I clinical trial of ATL-DC vaccination in conjunction with CSF-1R inhibitor (PLX3397) and PD-1 mAb (Pembrolizumab) blockade, and develop predictive immunological and imaging biomarkers.
Aim 3: To elucidate the immunotherapy-induced resistance mechanisms by which immuno-suppressive myeloid cells inhibit anti-tumor immune responses in pre-clinical animal models and in newly diagnosed glioblastoma patients.
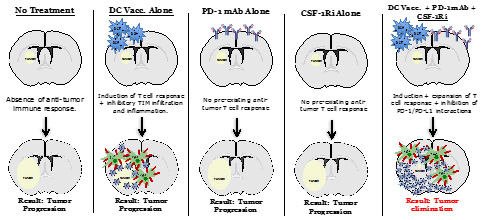
Figure for Project 1. Depiction of the critical immune components that regulate effective anti-tumor immune responses for malignant gliomas.
Project 2: Overcoming drug-induced resistance to intrinsic apoptosis in glioblastoma
Project Co-Leaders:
David Nathanson, PhD (Basic Science Leader)
Timothy Cloughesy, MD (Clinical Science Leader)
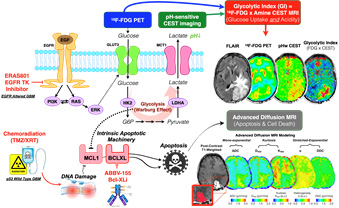
Glioblastoma (GBM) tumors are defined by high resistance to therapy-induced cell death. Through a combination of molecular and functional profiling of the intrinsic apoptotic machinery, we have identified that all GBM are comprised of two molecular intrinsic apoptotic blocks — MCL-1 and BCL-xL — to prevent tumor cell death. The goal of Project 2 is to translate rationally-designed drug combinations, consisting of new and pre-existing clinical agents, that selectively ablate the GBM dual apoptotic barrier to promote tumor cell death and induce durable clinical responses in patients.
Aim 1: To investigate whether a novel, antibody drug conjugate with a potent warhead against an essential apoptotic block has anti-tumor effects when combined with TMZ/radiation or a new, clinical brain penetrant EGFR TKI in pre-clinical GBM models.
Aim 2: To conduct a “window of opportunity” clinical trial to explore whether these new clinical drugs can ablate the two intrinsic apoptotic blocks in recurrent GBM patients.
Aim 3: To identify potential mechanisms of resistance to targeting the intrinsic apoptotic machinery in diverse pre-clinical GBM samples.
Together, the studies proposed in this application present a new therapeutic paradigm through specific manipulation of intrinsic apoptotic pathways in malignant glioma and have the long-term potential to shift current approaches in glioma therapy.
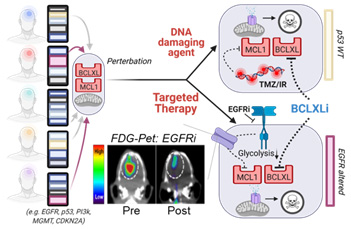
Figure for Project 2. All GBM have a dual apoptotic barrier — MCL1 and BCL-xL. Drug perturbations remove the MCL1 block in a genotype specific manner, creating an exclusive dependency on BCL-xL for survival. For EGFR TKI, changes in FDG uptake is a surrogate for ablation of MCL1
Project 3: Inhibition of radiation-induced phenotype conversion in glioblastoma
Project Co-Leaders:
Frank Pajonk, MD, PhD (Basic Science Leader)
Leia Nghiemphu, MD (Clinical Science Co-Leader)
Albert Lai, MD, PhD (Clinical Science Co-Leader)
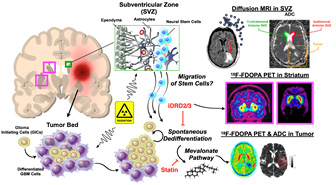
The overarching goal of Project 3 is to improve radiotherapy (RT) outcome for patients suffering from glioblastoma (GBM). In our last SPORE funding period, we used patient-derived glioblastoma cell lines, patient-derived orthotopic xenografts (PDOXs), and dopamine receptor antagonists (DRAs) to understand how glioma cells react to the combination of radiation and dopamine receptor inhibition. We found that the combination of RT with DRAs upregulated the fatty acid/cholesterol biosynthesis pathway, and that blocking this pathway led to markedly increased survival in tumor-bearing mice. We therefore hypothesize that blocking the radiation-induced phenotype conversion of non-stem GBM cells into radiation-resistant glioma-initiating cells using the dopamine receptor antagonist quetiapine (QTP) and statins improves radiation responses, generates an exploitable metabolic vulnerability, and can be safely applied in patients with recurrent glioblastoma.
Aim 1: To optimize the timing, sequence, and bioavailability of combination treatment with RT/QTP and statins that induces alterations in lipid metabolism and extends survival in tumor-bearing animal models
Aim 2: To test if RT/QTP induces changes in lipid metabolism that can lead to a vulnerability to be targeted for therapeutic gain in a first-in-human combination Phase I clinical trial of RT/QTP +/- statins
Aim 3: To determine the mechanisms by which RT/QTP-induced changes in lipid metabolism may be reversed by statins and prolong survival
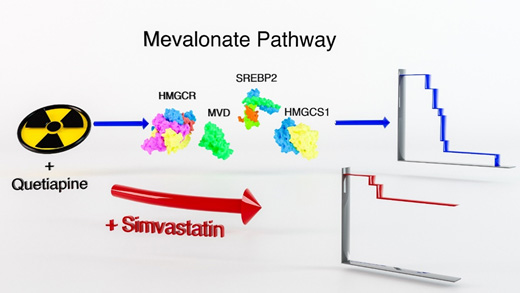
Figure for Project 3. Compared to radiation treatment alone, a combination of radiation and the dopamine receptor antagonist quetiapine significantly improves the median survival in mouse models of glioblastoma. At the same time, this combination treatment up-regulates gene expression of key enzymes in the mevalonate pathway with subsequent up-regulation of cholesterol biosynthesis. Targeting the rate-limiting enzyme in the mevalonate pathway, HMGCR (3-hydroxy-3-methylglutaryl-CoA reductase), using the statin simvatstain down-regulates cholesterol biosynthesis in glioma cells. A triple combination of radiation, quetiapine and simvastatin further improves median survival in mouse models of glioblastoma.
Administrative Core
Core Directors:
Linda M. Liau, MD, PhD, MBA (Core Director)
Timothy Cloughesy, MD (Core Co-Director)
Robert M. Prins, PhD (Core Co-Director)
The Administrative Core is responsible for the oversight and daily functions of the SPORE and provides organizational leadership for the overall successful conduct of the program. It provides organizational leadership and administrative support for all of the aspects of the SPORE, including coordination and communication between all investigators and committee members. It provides scientific management, including ongoing management and scientific review of all projects and cores to ensure that the stated scientific goals of the SPORE are met.
Biospecimen and Pathology Core (BiPC)
Core Directors:
Fausto J. Rodriguez, MD (Core Director)
Harley I. Kornblum, MD, PhD (Core Co-Director)
David A. Nathanson, PhD (Core Co-Director)
The Biospecimen and Pathology Core (BiPC), a biorepository and multifaceted research resource, provides critical support for the translational and diagnostic/therapeutic studies of the three major UCLA Brain SPORE projects, which are directed at understanding glioma biology and developing novel therapeutic approaches for brain cancer. In addition, the BiPC supports various developmental research projects and career enhancement programs. The SPORE projects have a critical need for high-quality brain tumor biospecimens, associated clinical and molecular annotations, histology, tissue microdissection, immunohistochemistry and characterization of models systems including neurospheres and xenografts. Neuropathology and biobanking expertise exist to accomplish the research aims and tightly interacts with the other cores.
Aim 1: To optimally collect, store, and distribute high-quality brain tumor biospecimens with detailed clinical and molecular annotations (tumor tissue, blood, urine, and cerebrospinal fluid) and implement biobanking best practices.
Aim 2: To perform molecular characterization of patient-derived tumor tissue and specific project models using histologic analysis, immunohistochemistry and genomic tools, including characterization of the tumor microenvironment and apoptotic machinery and single-cell RNA sequencing.
Aim 3: To create, provide, and characterize novel human biospecimen derivatives, gliomaspheres, patient-derived orthotopic xenografts (PDOX), and organoids that represent the molecular diversity of glioblastoma.
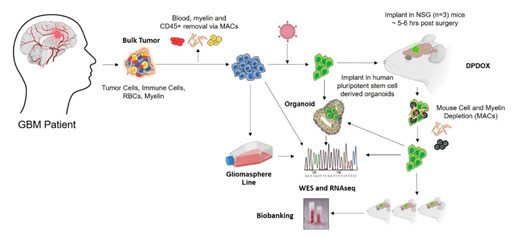
Figure for Biospecimen and Pathology Core. This figure shows the workflow of the Core with respect to tissue collection, and the development and characterization of relevant models.
Neuro-Imaging Core (NIC)
Core Director(s):
Benjamin M. Ellingson, PhD (Core Director)
Johannes Czernin, MD (Core Co-Director)
The goal of the Neuro-Imaging Core (NIC) is to provide advanced MRI and PET imaging support with established reliability and consistency to SPORE project investigators for their respective projects. The NIC uses quantitative μMRI and μPET for pre-clinical imaging (Aim 1), state-of-the-art MR and PET imaging acquisition, advanced post-processing, and novel analysis tools for clinical imaging of patients in novel clinical trials (Aim 2), and professional expertise and resources for traditional and enhanced radiographic response assessment for the clinical trials (Aim 3).
For Project 1, we theorize that tumors treated with DC vaccination and immune modulation will result in significantly higher uptake on immunoPET (89Zr-IAB22M2C Anti-CD8+ Minibody PET) and a significant decrease in tumor T2* on TAM-sensitive ferumoxytol MRI.
For Project 2, we theorize that a combined imaging biomarker — simultaneous 18F-FDG PET and pH-weighted CEST-EPI — may be a valuable imaging biomarker for glycolytic flux, which can be perturbed using EGFR inhibition (in EGFR altered GBM) or chemoradiation (in p53 wild type GBM).
For Project 3, we theorize that radiation therapy changes the microstructure of the subventricular zone (SVZ), known to harbor adult neural stem cells, as measured via diffusion MRI.
Biostatistics and Bioinformatics Core (BBC)
Core Directors:
Gang Li, PhD (Core Director)
Thomas Graeber, PhD (Core Co-Director)
The overarching goal of the Biostatistics and Bioinformatics Core of the UCLA Brain SPORE is to provide comprehensive support in the areas of biostatistics and bioinformatics for all research projects, developmental research and career enhancement programs, and other cores of the UCLA SPORE in Brain Cancer. This Core has two specific aims:
Aim 1: Biostatistics Support: Provide SPORE investigators broad-based statistical support for the individual SPORE projects. This includes statistical advice on study design, analysis and interpretation of experimental results, preparation of manuscripts, development and submission of new grant applications, data management, and the design, analysis, conduct, and monitoring of SPORE investigator-initiated clinical trials. Core 3 also develops relevant innovative methodologies and analytical tools that address specific needs from SPORE projects.
Aim 2: Bioinformatics Support: Provide infrastructure and bioinformatics leadership to design and carry out projects using high-throughput technologies, such as genomic, transcriptomic, and/or lipidomic data from tumors and/or immune cells, from bulk tissue and/or single cell next generation sequencing (NGS), and from patient tumors involved in clinical trials and from preclinical xenograft and immunocompetent mouse models. Up-to-date bioinformatics software and pipelines are provided.
Developmental Research Program
Program Director:
Robert M. Prins, PhD
The goal of the UCLA SPORE in Brain Cancer Developmental Research Program (DRP) is to foster the exploration of innovative research ideas through the funding of pilot developmental research projects in the field of brain cancer. This program will identify new, innovative projects for funding that will constantly renew the vigor of our Brain SPORE research portfolio and add to, augment, or replace current major research projects. We expect to select three highly innovative proposals for funding each year.
Career Enhancement Program
Program Director:
Linda M. Liau, MD, PhD, MBA
A central mission of the UCLA SPORE in Brain Cancer is to stimulate, recruit, develop, and retain new investigators in the field of brain tumor research, with particular focus on translation of scientific discoveries to novel clinical applications. The UCLA SPORE in Brain Cancer Career Enhancement Program (CEP) provides a research support resource that allows junior faculty to transition to independent investigator status with adequate peer-reviewed funding, and helps established investigators to focus or redirect their research to the area of brain cancer. Each year, we anticipate supporting two to three highly innovative candidates with CEP funds.







