The Memorial Sloan Kettering Specialized Program of Research Excellence (SPORE) in Leukemia
Memorial Sloan Kettering Cancer Center
Principal Investigator(s):

Omar Abdel-Wahab, MD

Martin Tallman, MD
- Principal Investigator(s) Contact Information
- Overview
- Project 1: Increasing therapeutic efficacy in isocitrate dehydrogenase (IDH)-mutant acute myeloid leukemia (AML)
- Project 2: Defining and exploiting genetic dependencies in complex karyotype AML
- Project 3: Therapeutic inhibition of splicing through inhibition of protein arginine methylation in leukemia
- Project 4: Chimeric Antigen Receptor T-Cell Therapy for the Treatment of Acute Myeloid Leukemia
- Administrative Core
- Biospecimen Core
- Genomics Core
- Biostatistics Core
- Developmental Research Program
- Career Enhancement Program
Principal Investigator(s) Contact Information
Omar Abdel-Wahab, MD
Director, MSK Center for Hematologic Malignancies,
Associate Attending, Leukemia Service
SKI, Human Oncology & Pathogenesis Program
Memorial Sloan Kettering (MSK)
Sloan Kettering Institute (SKI) for Cancer Research
1275 York Ave, Box 20
New York, NY 10065
(646) 888-2796
Martin Tallman, MD
Chief, Leukemia Service
Memorial Sloan Kettering Cancer Center
1275 York Ave
New York, NY 10065
(212) 639-3842
Overview
Despite recent progress, the majority of AML patients relapse following treatment, targeted therapeutic approaches for many recurrent molecular subtypes of AML are still lacking, the development of effective immunotherapeutic approaches for AML has been challenging, and survival rates in AML patients remain low. Although five-year overall survival rates for adults with AML have increased in the U.S. per decade since 1975, the overall five-year survival rate for adults with AML is 25-29%. To improve outcomes, there is an urgent need to develop mechanism-based, targeted, and immune-based therapies for AML patients and to identify novel biomarkers for risk classification and response to therapy.
To this end, the Memorial Sloan Kettering Cancer Center (MSK) Specialized Program of Research Excellence focuses on defining mechanisms that contribute to AML development and resistance to therapy, performing preclinical studies on novel molecular and immunologic targets in AML, and rapidly translating preclinical insights to innovative clinical trials for AML patients.
The overall translational aims of our SPORE program are to 1) interrogate genetic and molecular pathways required for AML initiation and maintenance; 2) develop novel targeted therapies and immunotherapeutic approaches for AML based on recurrent genomic alterations and leukemia stem-cell (LSC) specific markers; and 3) identify and validate the mechanism of action, therapeutic efficacy, and predictors of response/resistance of mechanism-based therapies for AML patients.
Our SPORE includes four projects, each addressing a different unmet need in the clinical management of AML:
Project 1. Increasing therapeutic efficacy in isocitrate dehydrogenase (IDH)-mutant AML
Project 2. Defining and exploiting genetic dependencies in complex karyotype AML
Project 3. Therapeutic inhibition of splicing through inhibition of protein arginine methylation in leukemia.
Project 4. Chimeric Antigen Receptor T-Cell Therapy for the Treatment of Acute Myeloid Leukemia.
Four shared resources support these projects: the Biospecimen Core, Biostatistics, Genomics and Bioinformatics Core, and an Administrative Core. This SPORE also supports a Career Enhancement Program to support junior investigators in translational leukemia research and a Developmental Research Program to support innovative translational research.
Project 1: Increasing therapeutic efficacy in isocitrate dehydrogenase (IDH)-mutant acute myeloid leukemia (AML)
Project Co-Leaders:
Ross Levine, MD (Basic Co-Leader)
Eytan Stein, MD (Clinical Co-Leader)
Although routine use of dose-intense chemotherapy regimens leads to clinical benefit in acute myeloid leukemia (AML), most patients subsequently relapse. As such there is a pressing need to develop novel therapies for AML patients based on insights into the molecular pathogenesis of different AML subtypes. We and others have genetically and functionally characterized the contribution of recurrent somatic alterations to AML pathogenesis, including IDH1/IDH2 mutations which can cause dysregulation of DNA (represented in graphic below). We have developed genetically accurate models of AML with IDH1/2 mutations in concert with co-occurring disease alleles, which have led to novel insights into their role in disease pathogenesis and allowed us to identify and credential IDH1/2 inhibitors as a therapeutic approach in AML. These studies have led to novel, mechanism-based clinical trials, and the first small molecule IDH1 (ivosidenib) and IDH2 (enasidenib) inhibitors are now approved for relapsed/refractory IDH1/2 mutant AML. These agents induce significant responses, including complete responses, in a subset of IDH-mutant AML patients, however not all patients respond to IDH1/2 inhibition and a subset of patients relapse following responses to IDH inhibition. Despite these important advances, there remain important questions relating to mechanisms of sensitivity and resistance to small molecule IDH1/2 inhibition in AML, and there is a need to increase therapeutic efficacy and further improve outcomes in this molecularly defined AML subset. First, although IDH1/2 inhibitors demonstrate efficacy in clinical studies, the role of co-mutated disease alleles in determining sensitivity to these agents has not been fully delineated. In addition, the mechanisms that can lead to therapeutic resistance to AML targeted therapies have not been defined. Therefore, we will use preclinical studies and analysis of primary samples from patients treated with IDH1/2 inhibitors to delineate molecular predictors of sensitivity and resistance to IDH inhibitors, and to test novel combination therapeutic approaches to increase therapeutic efficacy in IDH1/2-mutant AML. This will include mechanism-based clinical trials in genetically defined subsets.
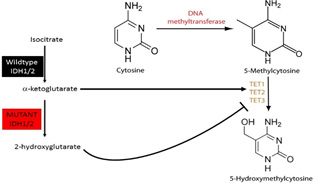
IDH1/2 mutations produce 2-HG which attenuates TET enzymatic function and dysregulates DNA methylation.
Project 2: Defining and exploiting genetic dependencies in complex karyotype AML
Project Co-Leaders:
Scott Lowe, PhD (Basic Co-Leader)
Martin Tallman, MD (Clinical Co-Leader)
Complex karyotype acute myeloid leukemia (CK AML) is defined by the presence of 3 or more detectable cytogenetic abnormalities and has one of the least favorable prognoses of any leukemia genotype (depicted below). Genomic characterization indicates that this disease lacks conventional druggable oncoproteins, but instead is characterized by a set of recurrent segmental deletions and mutations in the TP53 tumor suppressor gene, the latter of which are absent from normal karyotype AML and confer resistance to standard chemotherapies. To better characterize the pathogenesis of CK AML and develop new strategies to treat this disease, we will exhaustively analyze the genomes of a large cohort of CK AML samples using a new low-cost, high-resolution platform optimized in our group called “digital karyotyping”. These molecular features will be correlated with patient outcomes data, and used to generate murine and human models that accurately recapitulate CK AML-specific features. We will use these models to test a new therapeutic strategy for countering the pro-tumorigenic effects of p53 loss. This concept builds on preliminary data showing that p53 loss can perturb cellular metabolism in a manner that alters gene expression and drives aberrant self-renewal, and that reversing these effects with small molecule inhibitors can drive differentiation of p53-deficient AML. Specifically, we found that p53 mutations reduce levels of the metabolite aKG, producing similar effects of oncogenic IDH1/2 mutant proteins that have proven to be drug targets in other sub-types of AML. In models studied to date, the tumor suppressive effects of p53 are recapitulated by inhibiting the TCA enzyme 2-oxoglutarate dehydrogenase (OGDH) and, as such, we consider OGDH a prime candidate for validation and development in CK AML. Successful completion of this research will produce a detailed understanding of the genetic changes that accompany CK AML and allow for more faithful modeling of human disease. Validating OGDH as a novel drug target for CK AML will pave the way for clinical trials in this indication. Given the paucity of effective therapeutic options for patients with CK AML, the proposed studies address an urgent, unmet clinical need.
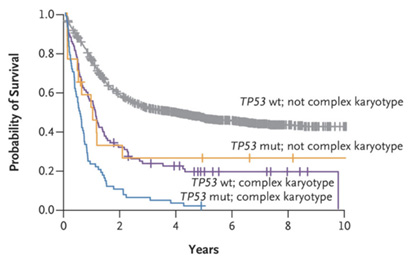
CK and TP53 mutations are associated with poor prognosis. Kaplan-Meier curves demonstrating reduced OS in AML featuring CK and/or TP53 mutation. Source: Papaemmanuil, E., et al. N Engl J Med. 2016.
Project 3: Therapeutic inhibition of splicing through inhibition of protein arginine methylation in leukemia
Project Co-Leaders:
Omar Abdel-Wahab, MD (Basic Co-Leader)
Andrew Kung, MD (Clinical Co-Leader)
Leukemias often display genetic alterations that result in dysregulation of the epigenome. To identify potential epigenetic vulnerabilities, we recently performed a paired in vitro and in vivo shRNA screen in a number of acute leukemia cell lines. The results showed that myeloid as well as lymphoid leukemia cells are preferentially dependent on protein arginine methyltransferase (PRMTs), a family of enzymes that dimethylate arginine residues of many proteins. Prior studies have identified PRMT5 as a promising therapeutic target in cancer, which has led to an ongoing phase I clinical trial of a PRMT5 inhibitor for patients with refractory solid tumors and Non-Hodgkin’s lymphoma. However, which substrates of PRMTs are most critical for anti-cancer effects of PRMT inhibition remains unknown and biomarkers predicting response to PRMT inhibition are greatly needed.
Toward understanding the anticancer effects of PRMT inhibitors, we have found that blocking PRMT function perturbs RNA splicing, and that inhibiting either symmetric (mediated by PRMT5) or asymmetric dimethyl arginine methylation (by Type I PRMTs) results in strong preferential killing of spliceosomal mutant leukemias over wild-type (WT) counterparts (depicted below). Moreover, we have observed synergistic effects of combining both type I with type II PRMT inhibition and/or inhibition of core spliceosome function. We therefore hypothesize that the main cytotoxic effect of PRMT inhibition results from modulation of splicing.
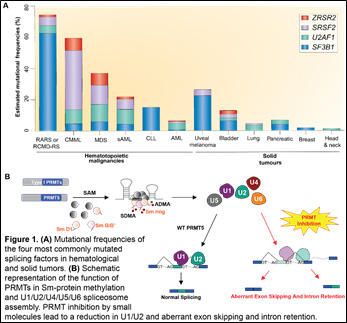
We are examining this by characterizing the effects of type I or type II PRMT inhibition on pre-mRNA splicing, gene expression, and the methyl-arginine proteome in WT or spliceosomal-mutant leukemia cells. We are also evaluating whether combining inhibitors of type I PRMTs, type II PRMTs, and the splicing factor SF3b enhances toxicity to myeloid and lymphoid leukemia cells, and the relationship between these inhibitors’ efficacy and mutations in various splicing factor genes. Finally, we are identifying biomarkers of efficacy of PRMT5 inhibition using samples from a phase I/II trial of GSK’s small molecule PRMT5 antagonist for the treatment of patients with refractory AML, CMML, and MDS. This project stands to greatly improve understanding of the molecular basis for the efficacy of PRMT inhibitors in cancer and to advance these drugs toward clinical trials in leukemia.
Project 4: Chimeric Antigen Receptor T-Cell Therapy for the Treatment of Acute Myeloid Leukemia
Project Co-Leaders:
Renier Brentjens, MD (Basic Co-Leader)
Anthony Daniyan, MD (Clinical Co-Leader)
There is an urgent and critical need for the development of leukemia stem cell (LSC)-directed therapeutic approaches for the treatment of acute myeloid leukemia (AML). One such strategy is targeting antigens that are specific to LSCs but absent from normal hematopoietic stem cells (HSCs). CD371 (CLEC12A, CLL-1), which is present on mature myeloid cells, has been described as one such targetable disease marker given its presence on both bulk AML cells and LSCs. Although not ubiquitously expressed on all AML cells, it is expressed in up to 95% of AML patients, is enriched on LSCs and chemoresistant AML subpopulations, and most importantly, is absent on HSCs.
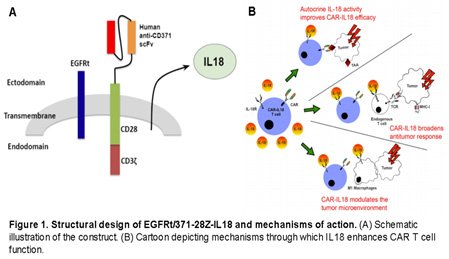
We have successfully developed and validated a fully-human CD371-targeted chimeric antigen receptor (CAR) T cell product which also secretes IL-18 (depicted below). Given that our CD371-targeting motif is entirely human, it is expected to have reduced immunogenicity and thus minimizes host-mediated CAR T cell-directed immune elimination in the context of constitutive IL18 secretion. In addition, IL18 secretion is predicted to enhance CAR T cell persistence and modulation the tumor microenvironment (TME) by increasing and activating immune cell infiltrates, leading to the induction of an endogenous T cell mediated anti-tumor immune-response capable of eradicating antigen-negative tumor cell subpopulations.
Our central hypothesis is that CD371-targeted IL18-secreting CAR T cells will lead to eradication of antigen-positive chemoresistant and LSC subpopulations, and the induction of an endogenous AML-reactive T cell response that will lead to elimination of antigen-negative disease, without long-term HSC toxicity. This will be tested in AML patient-derived xenograft models of heterogeneously antigen-positive disease and a phase I clinical trial with CD371-targeted IL18-secreting CAR T cells in patients with relapsed/refractory AML. This effort will therefore assess the safety and efficacy of this novel CAR-T cell approach in both preclinical and clinical settings, and will also address biomarkers of response and efficacy in numerous correlative studies.
Administrative Core
Core Directors:
Omar Abdel-Wahab, MD (Core Director)
Martin Tallman, MD (Core Co-Director)
The Administrative Core provides centralized scientific, managerial, and administrative oversight of the SPORE in Leukemia at Memorial Sloan Kettering Cancer Center. The core will leverage institutional resources and coordinate institutional interactions as well as administrative, fiscal, regulatory, and data management activities to facilitate efficient achievement of the translational research objectives. The Administrative Core will also include personnel trained in financial management, project management, grants administration, scientific and medical editing, data management, and administrative support who together will support the Core Director in achieving the SPORE program objectives.
Biospecimen Core
Core Director:
Elisa De Stanchina, PhD
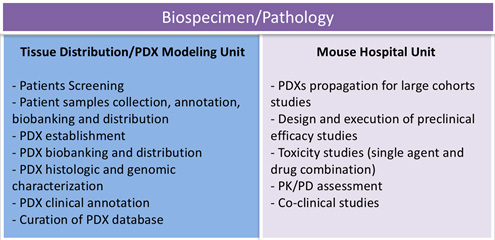
The mission of the Biospecimen Core will be to assist investigators with the development of preclinical models that accurately reflect the genomic landscape of human acute myeloid leukemia (AML), and their use in preclinical efficacy studies. The Core will play a central role in annotating, distributing, and tracking biospecimens from AML patients enrolled in biospecimen banking and therapeutic research protocols. Detailed biospecimen annotation, including documentation of pre-analytic processing variables, pathology findings, and patient clinical history will be recorded in robust relational databases. The Core will be composed of two highly integrated units: The Tissue Collection and PDX Modeling Unit will be responsible for providing access to human tissues and PDX models. The Mouse Hospital Unit will provide an integrated infrastructure to support pre- and co-clinical trials exploring efficacy of single agent and/or combinatorial treatments in relevant models using standardized protocols that mimic those used in patients. The MSK Mouse Hospital houses the only academic GLP-compliant facility in the region, providing investigators with the unique opportunity to perform GLP-compliant, IND-enabling safety toxicology studies for novel compounds and biologics in-house.
Genomics Core
Core Directors:
Elli Papaemmanuil, PhD (Core Co-Director)
Richard Koche, PhD (Core Co-Director)
The mission of the Genomics Core of the Memorial Sloan Kettering Cancer Center SPORE in Leukemia is to lead the analysis and interpretation of studies investigating the genetic, transcriptional, and epigenetic mechanisms contributing to AML development and resistance to therapy, and to identify potential therapeutic synergies and vulnerabilities through the integration of both targeted and unbiased genomic data. Achieving the transformative advances required for each Research Project to meet its goals will be greatly reliant on access to cutting-edge bioinformatics expertise. This expertise will be utilized to simultaneously focus a wide range of genome-wide data types on specific biological questions from each Project while also being broad enough to capture potentially shared mechanisms of response and resistance between the four Research Projects. Prior to the initiation of all studies, Core staff will consult with SPORE investigators to discuss the underlying scientific premise and translational goals of the project, to help the investigators select the ideal genomic techniques, sequencing parameters, and robust analytical methods.
Biostatistics Core
Core Directors:
Mithat Gönen, PhD (Core Director)
Sean Devlin, PhD (Core Co-Director)
The role of the Biostatistics Core is to support the investigators of the Leukemia SPORE in their research efforts, including laboratory experiments, molecular studies, and analysis of clinical trial and correlative data. Prior to the initiation of all studies, Core staff will consult with SPORE investigators to discuss the underlying scientific premise and translational goals of the project, to help the investigators select the most efficient and robust analytical methods, and to estimate sample sizes to ensure adequate power to address study objectives. In laboratory experiments, Core members will assist in the formulation of the experimental design and in the analysis and interpretation of the data at the conclusion of the study. For molecular studies using human tissues, Core members will closely interface with the members of the Genomics Core and will have primary responsibility for merging molecular and clinical data, and for performing appropriate statistical and bioinformatics analyses. The members of the Biostatistics Core will also work with SPORE investigators to format data for publication and assist with deposition of genomic data into public repositories.
Developmental Research Program
Program Directors:
Ross Levine, MD (Core Director)
Scott Lowe, PhD (Core Co-Director)
The majority of leukemia patients relapse after initial therapies, underscoring the need for improved targeted therapies and better predictors of clinical response. The Developmental Research Program (DRP) serve as an incubator for pilot projects that support the overall “bench-to-bedside and back” strategy of the program, supporting innovative pilot projects, technologies and research collaborations that have the potential to improve the diagnosis, prevention, and/or treatment of leukemia. The DRP will provide one to two years of funding to talented investigators or research teams from clinical and/or basic science backgrounds from the MSK community, neighboring institutions, or in support of collaborations with other Leukemia SPOREs. We will invite candidates annually to submit research proposals for peer-review by a committee of experts in leukemia and translational cancer research. We will prioritize research proposals that describe high-risk/high-reward hypothesis-driven work that has the potential to advance new avenues of leukemia investigation, or proposals that bring together exciting collaborative teams who will address ongoing problems in the leukemia field using new approaches and technologies.
Career Enhancement Program
Program Directors:
Omar Abdel-Wahab, MD (Core Director)
Martin Tallman, MD (Core Co-Director)
The MSK SPORE in Leukemia is dedicated to supporting the ongoing development of talented researchers who are pursuing independent translational research programs related to the diagnosis, prevention, or treatment of leukemia. The Career Enhancement Program (CEP) will provide research awards to support the scholarly development of 1) junior faculty from basic or clinical research backgrounds pursuing research related to the prevention, diagnosis and treatment of leukemia; 2) new or established faculty in other subject areas who wish to attain additional training and experience that will allow them to address areas of unmet need related to leukemia research; and 3) senior postdoctoral fellows who will continue their research program in leukemia and who are within one year of a faculty appointment. We will recruit up to two qualified investigators per year to receive CEP funding, with possibility of renewal for a second year.







