SPORE University of Texas MD Anderson Cancer Center-Leukemia
The University of Texas MD Anderson Cancer Center
Principal Investigator(s):

Marina Konopleva, MD, PhD

Elizabeth Shpall, MD
- Principal Investigator(s) Contact Information
- Overview
- Project 1: New Epigenetic Therapy Targets
- Project 2: Anti-PR1 Immune Therapy for Myeloid Leukemia
- Project 4: Off-the-shelf engineered cord blood-derived natural killer cells for the treatment acute lymphoblastic leukemia
- Project 5: Targeting Oxidative Phosphorylation in AML
- Administrative Core
- Pathology & Tissue Core
- Biostatistics, Data Management & Bioinformatics Core
- Developmental Research Program
- Career Enhancement Program
- Institutional SPORE Website
Principal Investigator(s) Contact Information
Marina Konopleva, MD, PhD
Professor/Director
The University of Texas MD Anderson Cancer Center
1400 Holcombe Blvd., Unit 428
Houston, TX 77030-4009
(713) 794-1628
Elizabeth Shpall, MD
Professor/Director
The University of Texas MD Anderson Cancer Center
1400 Holcombe Blvd., Unit 423
Houston, TX 77030-4009
(713) 745-2161
Overview
The M.D. Anderson Specialized Program of Research Excellence (SPORE) seeks to discover new therapies and actionable targets in leukemia and rapidly translate into major changes in the standard of care for patients with leukemia, through collaborative effort of Clinical Investigators, Biostatisticians, and Basic Investigators.
Our SPORE includes four translational research projects.
- Project 1: New epigenetic therapy targets.
- Project 2: Anti-PR1 Immune Therapy for Myeloid Leukemia.
- Project 4: Off-the-shelf engineered cord blood-derived natural killer cells for the treatment acute lymphoblastic leukemia.
- Project 5: Targeting Oxidative Phosphorylation in AML.
These projects are supported by three shared resources: Core 1. Administration Core; Core 2. Pathology & Tissue Core; and Core 3. Biostatistics, Data Management & Bioinformatics Core. This SPORE also supports a Career Enhancement Program to support junior investigators in translational leukemia research and a Developmental Research Program to support innovative translational research.
Project 1: New Epigenetic Therapy Targets
Project Co-Leaders:
Jean-Pierre Issa, MD (Basic Co-Leader)
Hagop Kantarjian, MD (Clinical Co-Leader)
Epigenetic therapy aims to reprogram gene expression in cancer cells to achieve a therapeutic effect. To date, DNMT inhibition is the most effective form of epigenetic therapy in myeloid leukemias. We have developed and validated a live cell assay to screen for drugs that achieve the same degree of epigenetic reprogramming as DNMT inhibition. Using this screen, we discovered a new class of epigenetic drugs that activate silenced expression through inhibition of CDK9. CDK9 is a transcriptional regulator previously linked to gene activation through the pTEFb complex that phosphorylates RNAPII and promotes transcriptional elongation. Our new data now place CDK9 at the heart of a node that regulates both gene silencing and activation in proliferating cells. As such, targeting CDK9 has pleotropic effects on gene expression that appear ideal from an anti-tumor perspective: One observes simultaneous gene activation (of tumor suppressors), repression (of oncogenes), and induction of an interferon immune signature, which may be immune-sensitizing. Known CDK9 inhibitors (flavopiridol, SNS-032) have activity in leukemias but are marred by serious chemotherapy-like toxicities. Examining published data, we find that doses of these drugs in use clinically are at least an order of magnitude higher than what is needed to inhibit CDK9, and we speculate that the toxicity observed is typical of cross-target inhibition of other CDKs (e.g. CDK1/2). Thus, we hypothesize that low doses of CDK9-selective drugs may preserve activity through epigenetic effects of CDK9 inhibition, while reducing toxicity by avoiding other CDKs. In this grant, we will elucidate mechanisms of epigenetic effects of CDK9, determine the downstream effects of CDK9 inhibition on cellular function and immune responses, and conduct a clinical trial of a new CDK9-selective drug in myeloid leukemias. Successful completion of these aims will introduce a new form of epigenetic therapy in the treatment of leukemias.
Project 2: Anti-PR1 Immune Therapy for Myeloid Leukemia
Project Co-Leaders:
Jeffrey Molldrem, MD (Basic Co-Leader)
Richard Champlin, MD (Clinical Co-Leader)
The development of novel immunotherapeutic approaches is critical in the fight against hematologic malignancies. With the goal of producing innovative, effective therapies against leukemia, we developed a humanized T cell receptor-like antibody, h8F4, which targets leukemia cells that express PR1/HLA-A2+. This first-in-class antibody has been approved for use in early phase clinical trials to treat patients with AML and/or MDS. We are actively collecting samples from patients being treated with h8F4 and performing informative studies. Our studies aim to characterize the AML blasts and leukemia stem cells under h8F4 therapy, to understand the relevant effects of the antibody and to explore novel mechanisms of action of h8F4 in vivo, including direct induction of apoptosis and potential ADCP activity. Studies will also examine alternative "next-generation" approaches to increase the potency of h8F4 through added mechanisms of action and to address resistance to h8F4 therapy, including 8F4 chimeric antigen receptor T cells (CAR-T) targeting PR1/HLA-A2, and anti-CD3/8F4 bi-specific monoclonal antibody constructs that enable CD3-positive T cells to recognize and eliminate PR1/HLA-A2-expressing leukemic cells.
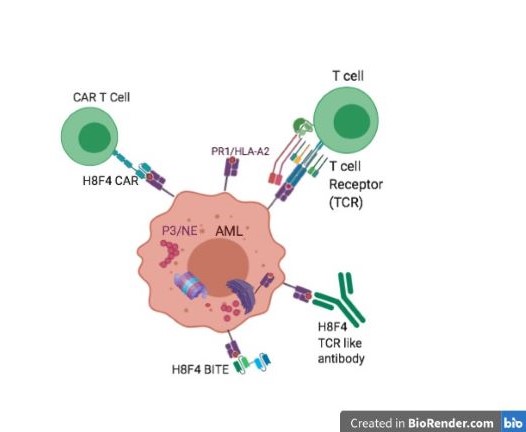
Figure 1. 8F4 bi-specific antibody, targets PR1/HLA-A2-expressing AML cells. Herrmann et al Front Immunol. 2019
Top left: 8F4 CAR T cells show anti-AML activity in vitro. Ma et al. Cytotherapy 2016
Top right: PR1 specific CTL have anti-leukemia activity Molldrem et al Blood 1997,
Bottom right: h8F4 - a monoclonal TCR-like antibody -targets PR1/HLA-A2 expressing AML cells. Sergeeva et al. Blood 2011, Leukemia 2016
Project 4: Off-the-shelf engineered cord blood-derived natural killer cells for the treatment acute lymphoblastic leukemia
Project Co-Leaders:
Katy Rezvani, MD, PhD (Basic Co-Leader)
Elizabeth J. Shpall, MD (Clinical Co-Leader)
Aim 1: Test the safety and antitumor activity of iC9/CAR.19/IL15-transduced CB-NK cells in patients with relapsed or refractory acute lymphoblastic leukemia.
NK-cells are attractive contenders for cell therapy as they exert potent antitumor cytotoxicity and unlike T-cells, do not cause graft versus host disease (GVHD) in the allogeneic setting. Through our Good Manufacturing Practice (GMP) facility, we have successfully engineered NK cells to express a specific CAR and are currently conducting a first-in-human trial of CAR.19/IL-15 transduced CB NK-cells in lymphoid tumors. Preliminary data prove their safety and efficacy with 8 of 11 patients with NHL or CLL treated to date achieving a complete or partial remission (NCT03056339). The data from the Phase I portion of this trial was published in the New England Journal of Medicine {Liu, 2020}. One of the problems with enrolling ALL patients was that most were too refractory to wait the 14 days for CAR-NK cell manufacture. We have now optimized the CAR-NK cell cryopreservation procedure so these will be off-the shelf products that will be infused immediately after thawing. This should markedly enhance accrual.
Aim 2: Track the fate of iC9/CAR.19/IL-15+ CB-derived NK cells after adoptive transfer and correlate the findings with disease response. Correlative studies have shown the persistence of the CAR-NK cells in the blood of the patients for up to one year following infusion.
Aim 3: Determine if targeting TGF-βR2, will further improve the therapeutic potential of IC9/CAR.19/IL-15+ CB-NK cells by protecting them from the immunosuppressive tumor microenvironment. The Rezvani laboratory developed and optimized a protocol for combined C9/CAR.19/IL-15 retroviral transduction and Cas9 ribonucleoprotein (Cas9 RNP)-mediated gene editing of TGF-βR2 to protect CAR-NK cells from TGF-β-mediated suppression in the TGF-β-rich ALL microenvironment. The gene-edited NK cells efficiently killed CD19 B cell cancer targets in vitro, even in the presence of exogenous TGF-β.
Project 5: Targeting Oxidative Phosphorylation in AML
Project Co-Leaders:
Giulio Draetta, MD (Basic Co-Leader)
Marina Konopleva, MD, PhD (Clinical Co-Leader)
Acute myeloid leukemia (AML) comprises a genetically and clinically heterogeneous group of aggressive hematological malignancies. Despite advances in molecular characterization of AML, the majority of patients will relapse and die of their disease. Studies by us and others showed that in AML, oxidative phosphorylation (OxPhos) generates intracellular energy and metabolic intermediates necessary to promote growth and support survival. Unlike normal hematopoietic stem cells, AML and leukemia stem/progenitor cells overexpress anti-apoptotic mitochondrial protein BCL-2, rely on OxPhos and are unable to utilize glycolysis when mitochondrial respiration is inhibited, indicating that the maintenance of mitochondrial function is essential for AML survival. We have identified a novel potent nanomolar inhibitor of OxPhos (OxPhosi) IACS-010759, found to inhibit complex I of OxPhos respiratory chain and block oxygen consumption. Inhibition of cellular respiration caused a metabolic catastrophe in subsets of AML and induces profound growth-inhibitory effects in primary CD34+ AML cells, with minimal toxicity in normal cells (Molina et al. Nat Med. 2018). IACS-010759 was well tolerated in mice, demonstrated strong efficacy in the in vivo xenograft studies utilizing the human AML patient-derived xenografts (PDX) and reduced phenotypically defined LSC fractions measured by CyTOF. We and others have further shown that AML cells including tumor-initiating stem/progenitor cells that survive chemotherapy or BCL-2 inhibition exhibit metabolic switch towards OxPhos dependency and that the anti-leukemia responses can be facilitated by targeting OxPhos in the setting of residual disease. These preclinical data prompt us to hypothesize that OxPhos inhibition constitutes a novel therapeutic approach that targets a unique metabolic vulnerability of AML including AML LSC; and that combined blockade of mitochondrial respiration by OxPhos and Bcl-2 inhibitors or chemotherapy will eliminate leukemia-initiating cells and produce objective responses. Pre-clinical studies in AML models will be conducted in parallel with a Phase 1 clinical trial. The overarching goal is to conduct a Phase 1/2 study of standard chemotherapy and BCL-2 inhibitor venetoclax combined with OxPhosi in patients with relapsed AML. These studies will facilitate mechanistic understanding of therapeutic vulnerability of AML and design of combination clinical trials targeting this unique metabolic reprogramming of energy-generating pathways.
Administrative Core
Core Directors:
Marina Konopleva, MD, PhD (Director)
Elizabeth J. Shpall, MD (Co-Director)
The Administrative Core provides the critical centralized administrative infrastructure to support and ensure the success of the SPORE. Core A provides essential, centralized administration to integrate the needs and synergize research potential among investigators representing various scientific disciplines. It oversees the conduct of proposed research and provides core services for a unified force for translational leukemia research.
Pathology & Tissue Core
Core Directors:
Steven Kornblau, MD (Director)
Carlos Bueso-Ramos, MD, PhD (Co-Director)
Jean-Pierre Issa, MD (Co-Director)
The major goal of the project is to provide patient derived material to researchers on the SPORE grant. The aims of the core are as follows:
Aim 1. Develop and maintain a repository of intact cells, serum, DNA, RNA and protein derived from blood and bone marrow specimens obtained from patients with AML at MD Anderson and Temple/Fox Chase Cancer Center.
Aim 2. Maintain a comprehensive, prospective interactive online database with detailed clinical and pathologic data for the samples received by Core 2.
Aim 3. Provide standard Reverse Phase Protein Arrays (RPPA) from AML patients to AML Project investigators.
Aim 4. Provide gene expression data interpretation for transcriptional profiling.
Aim 5. Facilitate inter- and extra- Leukemia Spore collaborations through sharing of blood and marrow resources.
Biostatistics, Data Management & Bioinformatics Core
Core Directors:
Xuelin Huang, PhD (Co-Director)
Xiaoping Su, PhD (Co-Director)
To serve all proposed SPORE projects, as well as the Career Enhancement and Developmental Research Programs, the Biostatistics and Bioinformatics Core has the following objectives:
- To provide the statistical design and sample size and power calculations for all projects.
- To provide bioinformatics data analysis of high-throughput and high-dimensional genomics data including whole genome/exome and transcriptome sequencing data.
- To facilitate prospective collection, entry, quality control, and integration of data for the basic science, pre-clinical, and clinical studies associated with the SPORE.
- To provide all statistical data analysis including descriptive statistical analysis, hypothesis testing, estimation, and modeling of prospectively generated data.
- To develop and adapt innovative statistical methods pertinent to biomarker-integrated translational leukemia studies.
- To generate statistical reports for all experiments within all projects.
- To collaborate and assist all project investigators in the publication of scientific results.
- To be a resource for intra- and inter-SPORE collaborations, including study design and developing databases for multi-center clinical trials.
Developmental Research Program
Program Directors:
Willian Plunkett, PhD
Marina Konopleva, MD, PhD
The goal of DRP program is to enable SPORE investigators to rapidly develop innovative research projects which could translate into clinical trials for leukemia patients, and could be developed into full projects and incorporated as a component of the Leukemia SPORE for competing renewal.
There are two to three awardees yearly.
Career Enhancement Program
Program Directors:
Marina Konopleva, MD, PhD
Willian Plunkett, PhD
CEP program supports promising young investigators at the junior faculty level who are interested in developing translational research in Leukemia. There are two to three awardees yearly, who may be either MD's or PhD's. Candidates are selected based on their previous track record, publications and their brief proposal that reflects their potential to pursue innovative translational research in leukemia.
Institutional SPORE Website
https://www.mdanderson.org/research/departments-labs-institutes/spores/leukemia-spore.html







