Dana-Farber/Harvard Cancer Center SPORE in Ovarian Cancer
Dana Farber Cancer Institute
Principal Investigators:

Alan D’Andrea, MD

Ursula Matulonis, MD

Cesar M. Castro, MD
- Principal Investigators Contact Information
- Overview
- Project 1: ATR inhibitor-mediated reversal of PARP inhibitor resistance in high-grade serous ovarian cancer (HGSOC)
- Project 2: Combined personal neoantigen-targeting cancer vaccines with immune checkpoint blockade for ovarian cancer
- Project 3: Evaluation of the Efficacy of Trametinib + Navitoclax in Recurrent Ovarian Carcinoma
- Administrative Core
- Pathology Core
- Biostatistics Core
- Organoids, Model Systems, and Biomarkers Core
- Developmental Research Program
- Career Enhancement Program
- Institutional SPORE Website
Principal Investigator(s) Contact Information
Alan D'Andrea, MD
Director, Susan F. Smith Center for Women’s Cancers
The Fuller-American Cancer Society Professor
Harvard Medical School
Dana-Farber Cancer Institute, HIM 243
450 Brookline Avenue
Boston, MA 02215
(617) 632-2112
Ursula Matulonis, MD
Chief, Division of Gynecologic Oncology
Brock-Wilson Family Chair
Dana-Farber Cancer Institute
Professor of Medicine
Harvard Medical School
Dana-Farber Cancer Institute
450 Brookline Ave.
Boston, MA 02115
(617) 632-2334
Cesar M. Castro, MD
Director of Gynecologic Oncology Program
Massachusetts General Cancer Center
55 Fruit Street
Boston MA
(617) 643-3778
Overview
The central focus of the Dana-Farber/Harvard Cancer Center (DF/HCC) SPORE in Ovarian Cancer is the understanding and overcoming mechanisms of primary and acquired treatment resistance in ovarian cancer. Our grant seeks to make translational progress in understanding resistance mechanisms that will lead to meaningful and improved outcomes of women with newly diagnosed ovarian cancer and those with relapsed ovarian cancer. The DF/HCC SPORE in Ovarian Cancer includes DF/HCC researchers from the Dana-Farber Cancer Institute, Massachusetts General Hospital (MGH), Brigham and Women’s Hospital, Beth Israel Deaconess Hospital, and Harvard Medical School (HMS). In addition, this SPORE includes investigators from outside of DF/HCC who are international experts in their fields, and they include researchers from the Massachusetts Institute of Technology (MIT) and the Broad Institute. We also have incorporated experienced patient advocates into this SPORE’s advisory structures.
Through the Ovarian Cancer SPORE of the DF/HCC, several of the most urgent questions in ovarian cancer therapy will be addressed. PARP inhibitors are now increasing being used to treat women with high grade serous ovarian cancer (HGSC) in both the newly diagnosed as well as recurrent settings. However, a rapidly emerging therapeutic problem for women with ovarian cancer is PARP inhibitor resistance and eventual progression through these agents. Accordingly, Project 1 involves clinical trials with drug combinations and correlative studies that have been designed to allow us to systematically extend the use of PARP inhibitors. We additionally recognize the emerging impact of immunotherapy on solid tumor treatment but also understand the limitations of immune checkpoint blockade as single agents for recurrence or added to chemotherapy for both newly diagnosed and recurrent ovarian cancer patients. In Project 2, we have designed a novel neoantigen vaccine trial for ovarian cancer patients. Third, HGSC patients with primary refractory disease or those patients with recurrence whose cancers harbor underlying RAS mutations such as low grade serous or mucinous cancers pose a particularly difficult clinical problem. Thus, in Project 3, we will explore novel non-platinum drug combinations, such as the combination of a BCL inhibitor and a MEK inhibitor and will examine predictive biomarkers and tumor responses evident in the extracellular activity. Our SPORE in Ovarian Cancer additionally has a Developmental Research Program (DRP) and a Career Enhancement Program (CEP) as well as 4 cores: Administrative, Pathology, Biostatistics, and Organoids, Model Systems, and Biomarkers. These cores will provide infrastructure and support for each of the Projects and any DRP and CEP projects.
Project 1: ATR inhibitor-mediated reversal of PARP inhibitor resistance in high-grade serous ovarian cancer (HGSOC)
Project Co-Leaders:
Geoffrey Shapiro, MD, PhD
Cesar M. Castro, MD
Lee Zou, PhD
More than half of high-grade serous ovarian cancers (HGSOCs) carry defects in homologous recombination (HR) repair, most commonly related to BRCA1 and BRCA2 alterations. Such alterations also compromise the stability of stalled replication forks in ovarian cancer cells. These features translate to sensitivity to poly (ADP-ribose) polymerase (PARP) inhibition so that the introduction of PARP inhibitors has transformed the treatment paradigm for this disease. These agents are now approved for all stages of ovarian cancer treatment: newly diagnosed patients and those with recurrent cancer as well. Unfortunately, acquired resistance has emerged as a high unmet clinical need. Current data suggest that the two major mechanisms of PARP inhibitor resistance include (1) restoration of HR; and (2) replication fork stabilization. The work of Project 1 investigators has shown co-occurrence of these mechanisms in ovarian cancer cell line models of acquired PARP inhibitor resistance. Consequently, strategies that address both HR restoration and replication fork stability represent promising avenues for development. Our preliminary data in cell lines suggest that inhibition of Ataxia telangiectasia and Rad3-related (ATR) or checkpoint kinase 1 (CHK1) is capable of reversing both mechanisms. Here, we will confirm these results in primary ovarian organoid cultures, as well as in BRCA-mutated patient-derived xenograft (PDX) models with acquired PARP inhibitor resistance, followed by translation to a clinical trial of combined ATR and PARP inhibition.
In addition to the problem of acquired resistance in initially responsive tumors, subsets of HGSOC demonstrating a high degree of replication stress, such as those harboring CCNE1 amplification, are refractory to PARP inhibition but sensitive to ATR or CHK1 inhibition. We have completed a trial randomizing gemcitabine to gemcitabine combined with ATR inhibition in platinum-resistant HGSOC (NCT02595892) that demonstrated superiority of the combination, particularly in patients with a platinum-free interval < 3 months. We hypothesize that the gemcitabine/ATR inhibitor combination may be particularly useful in HGSOCs under high replication stress. We will test this hypothesis in both preclinical models and in biopsy samples procured during the trial, with the development of biomarkers that may define HGSOCs with replication stress that predict sensitivity to ATR or CHK1 inhibition alone or in combination with gemcitabine.
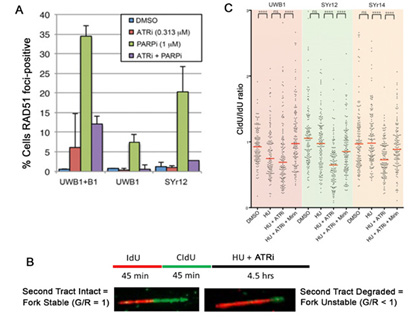
ATR inhibition reverses major mechanisms of PARP inhibitor resistance. (A) HGSOC UWB1 BRCA1-mutant HGSOC cells with acquired resistance by virtue of BRCA1-add-back (B1) or with acquired resistance after chronic exposure to olaparib (SYr12) have restored HR, assayed by RAD51 focus formation in response to PARP inhibition. This is reversed in the presence of an ATR inhibitor. (B) DNA fiber assay for assessment of replication fork stability. (C) Compared with UWB1 parental cells, cells with acquired PARP inhibitor resistance (SYr12 and SYr14) have stabilized replication forks. Fork stabilization is reversed in the presence of an ATR inhibitor. From Yazinski et al, Genes Dev, 2017.
Project 2: Combined Personal Neoantigen-Targeting Cancer Vaccines with Immune Checkpoint Blockade for Ovarian Cancer
Project Co-Leaders:
Panagiotis A. Konstantinopoulos MD, PhD
Catherine Wu, MD
Recent studies of immune checkpoint blockade (CPB) therapy have demonstrated its potent clinical activity across multiple tumor types but only modest effectiveness in ovarian cancer has been observed. Hence, the development of strategies to improve efficacy of CPB while minimizing immune-related toxicity is currently a high priority for ovarian cancer treatment. In parallel, large-scale cancer sequencing efforts have revealed the mutational landscape of tumors, which together with the maturation of predictive algorithms for binding of peptides to class I HLA molecules, have enabled the ability to systematically identify immunogenic epitopes arising from somatic mutations, called neoantigens. Unlike native proteins, neoantigens are not subject to the immune-dampening effects of central tolerance and yet exhibit exquisite tumor-specific expression. We and others have spearheaded efforts demonstrating the feasibility of using such computational tools to identify immunogenic candidate patient-specific mutated epitopes that are capable of stimulating tumor-specific T cell responses. This promising activity has led us to prospectively test the targeting of personal neopeptides as cancer vaccines. Recently, we demonstrated the proof-of-concept of the safety, feasibility and immunologic activity of immunizing patients with advanced melanoma and glioblastoma with personal vaccines consisting of up to 20 mutated epitopes per patient, delivered as synthetic long peptides (20-30mers) admixed with the potent immune adjuvant poly-ICLC, a TLR3 agonist (the combination of the neoantigen vaccine and poly-ICLC is termed Ott et al. Nature, 2017 and Keskin et al. Nature 2019. Of note, the induced neoantigen-specific T cell responses could recognize autologous tumor cells.
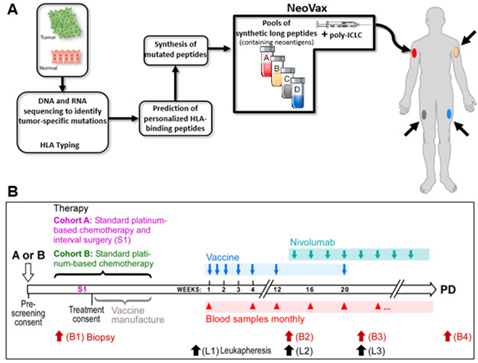
These results suggest that a promising treatment approach is the administration of cancer vaccines in combination with CPB therapy to enhance the effectiveness of CPB therapy. Using previously validated pipelines of neoantigen prediction, we identified at least 30 predicted neoantigens in approximately 70% of epithelial ovarian cancers of high grade serous histology and that higher number of predicted neoantigens is associated with improved overall survival in these tumors Strickland et al. Oncotarget 2016. We hypothesize that applying this approach to patients with EOC will effectively expand existing tumor-reactive T-cells and broaden the T-cell repertoire to include new tumor-specific T-cells thereby generating highly specific anti-tumor immunity with fewer autoimmune side effects. Neovax production is shown in Figure A; advances in prediction algorithms generated by our team now provide opportunities for studying the feasibility of generating neoantigen vaccines in tumors with intermediate mutation load, such as EOC, and for the testing of how the vaccine can be administered in conjunction with standard-of-care therapy Figure B. Epithelial ovarian cancer in the setting of low burden residual disease is an ideal setting for testing the biologic impact of combined personal cancer vaccine with CPB because we will have the ability to detect changes in both tumor-infiltrating immune cells and the tumor itself at site of disease in relationship to therapy and clinical response, through the direct analysis of patient tumor specimens collected through the course of treatment.
Project 3: Evaluation of the Efficacy of Trametinib + Navitoclax in Recurrent Ovarian Carcinoma
Project Co-Leaders:
Joan S. Brugge, PhD
Gad Getz, PhD
Ursula Matulonis, MD
The overall objective of this project is to investigate the clinical efficacy of a combination therapy targeting the RAS-ERK pathway serine/threonine kinase MEK and two anti-apoptotic proteins, BCL-2 and BCL-XL in ovarian cancer. Epithelial ovarian cancer is comprised of several subtypes, the most common being high grade serous carcinoma (HGSC). While initially sensitive to platinum drugs and poly ADP ribose polymerase (PARP) inhibitors, HGSC becomes increasingly therapy resistant. Additionally, other ovarian cancer subtypes, such as low grade serous (LGSC) and mucinous ovarian cancers, many of which are RAS/RAF mutated, are often platinum refractory, and new treatment strategies are needed. We hypothesize that combined MEK and BCL-2/XL inhibition will be active in refractory/relapsed ovarian cancer based on: (i) evidence that the RAS-ERK pathway is activated in a large percentage of ovarian cancers, (ii) the efficacy of combined MEK and BCL-2/XL inhibition in preclinical PDX models of chemoresistant HGSC, (2) preclinical models of RAS mutant tumors, and (iii) a Phase I clinical trial showing safety and tolerability of combined treatment with the MEK inhibitor trametinib and BCL-2/XL inhibitor navitoclax.
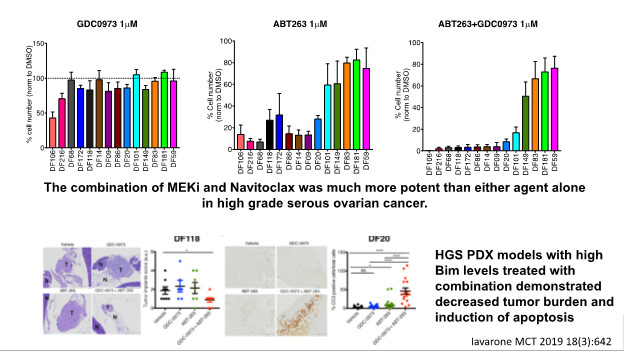
To test this hypothesis, we will carry out a Phase II clinical trial of trametinib plus navitoclax in recurrent platinum-refractory/resistant (1) HGSOC and low grade serous cancer (RAS wild type) and (2) RAS/RAF mutant ovarian cancers. In addition, we will investigate genetic and proteomic markers that correlate with efficacy. One focus will be on the protein BIM, which has been shown to correlate with efficacy to trametinib and navitoclax in HGSC PDX models and other cancers. We will also perform pre-clinical studies to identify therapeutic approaches that enhance the efficacy of trametinib plus navitoclax. Overall, the research in Project 3 will provide information critical to the identification of new therapies for hard-to-treat ovarian cancers resistant to current approaches.
Administrative Core
Core Directors:
Alan D’Andrea, MD
Ursula Matulonis, MD
Cesar M. Castro, MD
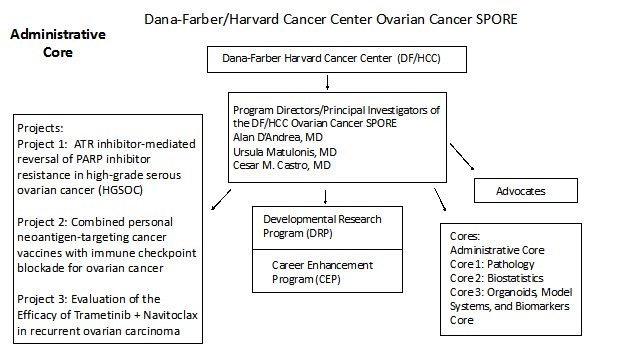
The goals of this Administrative Core are to provide leadership and oversight of this Ovarian Cancer SPORE grant, facilitate integration and collaboration of this Ovarian Cancer SPORE within the DF/HCC as well as with other SPORE grants and enable the success of the overall Ovarian Cancer SPORE and its individual components. The administrative core will coordinate and evaluate each of the components of the Ovarian Cancer DF/HCC SPORE grant in addition to providing leadership and operational oversight of all of the scientific, administrative, and fiscal aspects of the SPORE grant. The Administrative Core will facilitate scientific interactions within the SPORE, both within as well as outside of DF/HCC and assuring that findings are shared from ongoing research within the SPORE Projects. The Core also will also provide for engaging our advocacy community both within DF/HCC as well as outside of Harvard and to in addition, leverage the SPORE resources, scientific findings, and collaborations to generate additional funding for ovarian cancer research both within DF/HCC and outside as well. The SPORE PIs/PDs will equally provide fiscal and resource oversight to the projects and cores assuring translational research and clinical trial success. The administrative Core will ascertain that each project has adequate budgetary resources for success. The structure of the DF/HCC SPORE for Ovarian Cancer is detailed in this figure below:
Pathology Core
Core Directors:
David L. Kolin, MD, PhD
Sarah J. Hill, MD, PhD
Bo Rueda, PhD
Esther Oliva, MD
The integrated Pathology Core brings together experts in ovarian cancer pathobiology with the SPORE Project Investigators, the Organoids, Model Systems, and Biomarkers Core, and the DRP and CEP. The effectiveness of the Pathology Core lies in the efficient use of both separate and shared resources. The Pathology Core will be in close proximity to the investigators who require their services for the DF/HCC SPORE in Ovarian Cancer projects at their respective institutions. A centralized working group of pathologists from the participating institutions is the best way to ensure that this research will be properly carried out for the following reasons: 1) Efficient distribution of consented tissues is paramount because collected samples could find their way into multiple studies. Careful coordination with pathology, the consenting teams, and all research teams will ensure that a single consented case can be allocated to multiple researchers at different institutions. 2) Studies will be conducted largely at the two major institutions Brigham and Women’s Hospital/Dana Farber Harvard Cancer Center (BWH/DFHCC) and Massachusetts General Hospital (MGH), and it is likely that projects from multiple studies will be trying to access tissue from the same cases. The pathology core will coordinate tissue collection to ensure that the maximum amount of research can be done. 3) A central group of pathologists across all projects will allow for immediate and uniform tissue processing within each project and uniform interpretation of all data by a single group of trained experts.
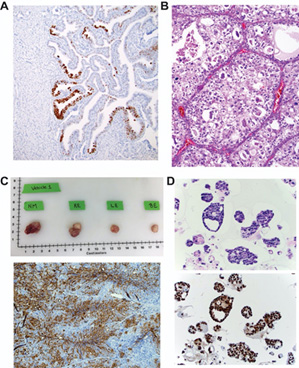
Pathology Core Expertise: The pathology core is led a by a group of clinicians and scientists from BWH, MGH, and DFCI with a long history of utilizing human tissue in translational and basic science research. Each brings general expertise in this area to aid all other aspects of the SPORE and has areas of specific expertise including A) expertise in studying ovarian carcinogenesis (Dr. Crum) shown here with a p53 stain demonstrating a p53 signature in a Li Fraumeni fallopian tube, B) expertise in the pathobiology of clear cell ovarian carcinoma (Dr. Oliva) shown here with an H&E image of a clear cell carcinoma, C) expertise in the study of mechanisms of treatment induced resistance in ovarian cancer using mouse in vitro and in vivo models of ovarian cancer (Dr. Rueda). Shown above are gross (top) and IHC (bottom) images of ovarian tumors from a PDX model post treatment from his lab, and D) expertise in organoid modeling of ovarian cancer to study mechanisms of drug response (Dr. Hill) shown here with an H&E (top) and p53 stain (bottom) of sections of high grade serous ovarian cancer organoids from her lab.
Biostatistics Core
Core Directors:
Steven Skates, PhD (MGH)
Nabihah Tayob, PhD (DFCI)
The DF/HCC Ovarian Cancer SPORE Biostatistics Core will collaborate on all research activities within the SPORE, including SPORE Projects, Developmental Research and Career Development projects, and other SPORE Cores, assure the highest standards of scientific rigor in areas of study design, statistical modeling, statistical inference, and data integrity. The specific aims of the Biostatistical Core are:
Specific Aims
Aim 1: To collaborate on SPORE Projects, Developmental and Career Enhancement Projects, and other Cores for study design, conduct, monitoring, statistical analysis, and reporting of translational, clinical and associated correlative studies, laboratory, and animal studies.
Aim 2: To collaborate with SPORE investigators on integrating laboratory analyses with endpoints from clinical studies and translational research through implementing data acquisition, data integrity, and data storage with statistical software.
Aim 3: To collaborate on short-term projects with the entire group of SPORE researchers, including Project Leaders and Core Leaders.
Aim 4: To collaborate on the evaluation of Developmental Research and Career Enhancement Programs through judgment on biostatistical design and analyses during the development and review of these projects.
Aim 5: To collaborate with SPORE investigators to identify experimental study design issues and biostatistical modeling arising in ovarian cancer research for which no standard methodologies are available and to encourage faculty associated with the SPORE and within the wider community of statisticians and bioinformaticists at DF/HCC to conduct research in developing new methods to address those problems.
Dr. Steven Skates and Dr. Nabihah Tayob are biostatisticians who have the expertise for the planning, study design, statistical modeling, analyses, and reporting of translational, clinical and associated correlative studies, epidemiological, and laboratory studies arising in all SPORE Projects, Developmental Research and Career Development projects, and other SPORE Cores. They have command of a wide range of statistical software required for implementing advance statistical modeling to meet challenges encountered when collaborating throughout the SPORE. This includes ensuring data integrity through innovative software for data collection, storage, and transfer, including coordination of laboratory results with outcomes from clinical studies and translational research. Centralizing expertise in the Biostatistics Core is an effective strategy to guarantee a high degree of integration among projects, which have interrelated analytic goals and needs.
Organoids, Model Systems, and Biomarkers Core
Core Directors:
Alan D’Andrea, MD
Neil Horowitz, MD
Joyce Liu, MD
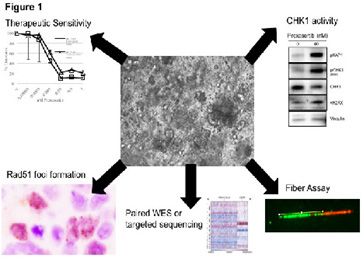
The overall purpose of the Organoids, Model Systems, and Biomarkers Core is to procure relevant clinical samples and develop assays and organoid models (see Figure 1) that will support the investigation and development of strategies to overcome primary and acquired resistance. Our first priority is to maintain and maximize access to consented tissues from women who are newly diagnosed with ovarian cancer or have residual or recurrent disease following chemotherapy. The second is to work with the investigators and the bank to ensure that project specific tumors that have been targeted and stored are successfully sequestered and retrieved on demand. The third is to ensure that the quality of the tissue is maximized and established by histologic review. Two tissue bank repositories are in place including the BWH/DFHCC (Drs. Liu, Matulonis) and MGH (Dr. Rueda) repositories. These repositories require multi-disciplinary co-operation in order to obtain, transport, and store tumor samples. Each institute has their own system set up for the retrieval and storage, but, the tissue bank staff from both institutions now meets on a monthly basis to maximize research and troubleshoot any processing or dispersal issues.
Developmental Research Program
Program Directors:
Ursula Matulonis, MD
Kevin Elias, MD
Alan D’Andrea, MD
The goals of the Developmental Research Program (DRP) of our SPORE in Ovarian Cancer are to successfully identify, fund, monitor, and support high risk/high potential pilot projects that take maximum advantage of new research opportunities within the Dana-Farber/Harvard Cancer Center Ovarian Cancer SPORE and to maintain this Program throughout the entire grant term. The success of this Ovarian Cancer SPORE will be partly dependent upon our ability to engage researchers within and outside of the DF/HCC community who can contribute novel ideas to the diagnosis and treatment of ovarian cancer. We are interested in attracting qualified candidates or groups of candidates who are at the academic level of Instructor level or above including scientist teams as well all levels of investigators. The DRP will be uniquely open to submissions from more senior investigators whose proposal involves the introduction of ovarian cancer targeting to established research programs. However, investigators at any stage will be encouraged to apply. Priority will be based upon the novelty of their proposals, evidence to support that the project can be completed within specified timelines and budget proposal, and a proven track record of accomplishment and productivity by the investigator. Those projects that have demonstrated great progress and translational research potential will have the potential to become full SPORE Projects but only with the approval of the DF/HCC Ovarian Cancer DRP Executive Committee. New and novel research ideas to better treat ovarian cancer will be greatly beneficial to current and future ovarian cancer patients.
Career Enhancement Program
Program Directors:
Cesar M. Castro, MD
Kevin Elias, MD
Alan D’Andrea, MD
The overall objective of the Career Advancement Program (CEP) is to increase the breadth and depth of the ovarian cancer translational investigator pool at the DF/HCC and across the US. We expect to train individuals who will become National / International leaders in the pathobiology, diagnosis, treatment and control of Ovarian Cancers. The first Specific Aim of the CEP is to identify appropriate candidates, primarily junior faculty members seeking to become independent investigators in ovarian cancer research, provide and secure funding for these investigators, and assure SPORE resources for awardees. We will focus on the rich applicant pool within the DF/HCC to recruit, train and retain the very best new talent in oncology. We are particularly committed to increase the diversity of the peer reviewed translational science investigators in ovarian cancer. The second Specific Aim is to promote a mentoring environment and plan that will assist the career enhancement and success of the CEP awardees. Through the environment in the DF/HCC, we will create individualized training programs for every funded junior faculty person and build a team of scientific, clinical and personal mentors around each one. The third Specific Aim is to monitor the scientific and academic progress of the awardees and the program, and every trainee will receive intensive support from Drs Castro, Matulonis, and D’Andrea, the program leaders and their own dedicated lab heads.







