Case Western Reserve University GI Cancers SPORE
Case Western Reserve University
Principal Investigator(s):

Sandford Markowitz, MD, PhD
- Principal Investigator(s) Contact Information
- Overview
- Project 1: Targeting 15-PGDH in Colon Cancer Risk, Prevention and Treatment
- Project 2: Translational Significance of a Mutational Signature of African American Colon Cancers
- Project 3: Molecular Markers for Early Detection of Esophageal Carcinogenesis
- Project 4: Targeting Glutamine Metabolism in Colorectal Cancer Harboring PIK3CA Mutations
- Administrative Core
- Biospecimen Core
- Biostatistics and Informatics Core
- Developmental Research Program
- Career Enhancement Program
Principal Investigator(s) Contact Information
Sanford D. Markowitz, MD, PhD
Principal Investigator
Ingalls Professor of Cancer Genetics
Case Comprehensive Cancer Center
Departments of Medicine, Genetics, and Molecular and Microbiology
Case Western Reserve University School of Medicine
Room 3-128
Wolstein Research Building
Case Western Reserve University
10900 Euclid Avenue
Cleveland, OH 44106-7285
(216) 368-1976
Overview
This proposal provides for a cutting edge Specialized Program in Research Excellence in gastrointestinal malignancies. This proposal places an emphasis on colorectal cancers (CRC), which are the second leading cause of cancer death in the United States and the industrialized world, with 134,000 new cases and 49,000 deaths in 2016 in the U.S. alone. In recognition of the extraordinary burden of mortality and morbidity caused by this disease, this proposed GI SPORE puts forward three cutting edge translational research projects that deal with new approaches for reducing deaths from colon cancer (SPORE Projects 1, 2, 4). SPORE Project 1 focuses on developing new strategies for CRC prevention through targeting the 15-PGDH tumor suppressor pathway discovered by the SPORE investigators. SPORE Project 2 targets elucidating the biological basis for colon cancer disparities among African Americans, based on the innovative discovery by the SPORE investigators of somatic mutations that uniquely target cancers arising in this population. SPORE Project 4 targets developing new therapeutic approaches for colon cancers with PIK3CA mutations, based on the project investigator's novel finding that these cancers are addicted to glutamine. In addition to this emphasis on colon cancer, this GI SPORE puts forward a fourth translational research project (SPORE Project 3) that focuses on molecular early detection of Adenocarcinoma of the Esophagus and on its biological precursor lesion, Barrett's Esophagus (BE). This reflects that esophageal adenocarcinomas (EAC) are the fastest growing cause of mortality from solid tumors in the U.S., with a 5-fold increase in incidence over the last 3 decades and a five-year survival rate of only 17%. Moreover, whereas cancer of the esophagus in the U.S. were once mostly squamous cancer of the upper two-thirds of the esophagus, now the large majority of are adenocarcinomas that arise in the lower one-third of the esophagus and develop in a background of histological change of BE. SPORE Project 3 now proposes a novel non-endoscopic biomarker-based approach for screening and early detection of Barrett's esophagus, the marker of the population at risk for EAC. The approach is based on the discovery by the SPORE Project investigators of a robust methylated DNA biomarker of BE coupled with their invention of a novel swallowable balloon device that enables obtaining esophageal brushings without requiring endoscopy.
The common theme of this GI SPORE is the application of cutting edge molecular approaches to develop new approaches for risk stratification, prevention, and treatment of gastrointestinal cancers. The proposal reflects the long history of collaborative studies by an investigator group that has been working closely together throughout the prior SPORE funding period, and well before. The aims of this SPORE sponsored research are:
Aim 1 (Project 1): To develop the 15-PGDH colon cancer suppressor gene, and its partner genes in regulating colon PGE2, as a new molecular pathway for identifying individuals at high CRC risk; for identifying individuals who will are sensitive (versus resistant) to CRC chemoprevention with aspirin; and for identifying novel CRC prevention strategies — via elucidating lifestyle and CRC risk factors that act to modulate this pathway.
Aim 2 (Project 2): To interrogate a molecularly defined sub-type of colon cancer that uniquely arises in African American individuals; to elucidate the biological basis for the development of the mutations that define these cancers; and to determine the impact this molecularly defined cancer subtype has on the disparities in colon cancer outcomes that are associated with African Americans.
Aim 3 (Project 3): To develop a non-endoscopic biomarker based approach for screening/early detection for BE; to test this approach in a human clinical trial; to further develop non-endoscopic biomarker based approaches for surveillance of BE for early detection of progression of BE to high grade dysplasia or early EAC; and to interrogate the functional role these biomarkers play in esophageal neoplasia.
Aim 4 (Project 4): To elucidate the mechanism by which CB-839 augments the activity of 5-FU in PIK3CA mutant CRC; to perform a human clinical trial testing responses of PIK3CA mutant colon cancers to 5-FU combined with CB-839, a glutamine metabolism inhibitor that targets glutaminase; to develop even more potent inhibitors of glutamine metabolism by developing inhibitors of glutamic pyruvate transaminase 2.
Aim 5: To foster development of new investigators and new approaches to translational research in GI Cancers through support of focused Developmental Research and Career Enhancement Programs.
Aim 6: To provide translational Investigators in G.I. cancers support from core resources that provide access to state of the art Biospecimens and Experimental Pathology, Statistical Methods, and Administrative Services.
Project 1: Targeting 15-PGDH in Colon Cancer Risk, Prevention and Treatment
Project Co-Leaders:
Sanford D. Markowitz, MD, PhD
L. Li, MD, PhD
N. Berger, MD
The premise of this proposal is that colon inflammation, as mediated by colon PGE2, is a key determinant of i) the risk of colorectal cancer (CRC) and ii) the efficacy of CRC prevention by aspirin. This premise reflects our team's seminal work identifying 15-PGDH as i) a key in vivo regulator of colon PGE2, ii) a biomarker of CRC risk, and iii) a predictive biomarker of aspirin's efficacy. We propose that a signature that now captures 6 key PGE2 regulatory genes will yield an even more powerful biomarker for improving CRC prevention, by enabling personalized strategies to improve risk-based screening and aspirin chemoprevention.
Colorectal cancer remains the second leading cause of cancer deaths in the United States. Colonoscopy screening has been a mainstay for reducing colorectal cancer deaths via early detection of colorectal cancers and adenomas. However, colonoscopy screening is generally recommended only once every ten years, and in the absence of biomarkers to identify individuals at high risk, development of interval cancers between colonoscopies remains a recognized challenge. PGE2 is a bioactive lipid metabolite that plays a key role in promoting colon neoplasia growth and development. Aspirin lowers PGE2, via inhibiting the COX enzymes that are required for PGE2 synthesis, and using aspirin reduces CRC incidence by roughly one-third. To meet the continuing challenge of persistently high colorectal cancer deaths, the US Preventive Services Task Force in 2016 endorsed aspirin use for primary prevention of colorectal cancer. However, reflecting uncertainty over which individuals benefit from aspirin chemoprevention, this endorsement was restricted only to individuals who meet criteria for using aspirin to also reduce cardiovascular risk, thus leaving a critical gap in knowing how to apply aspirin for reducing CRC cancer deaths in the general population, a challenge whose importance NCI leadership itself has highlighted.
Although PGE2 is a key driver of colon neoplasia, it is metabolically labile, and can only be studied in tissues specially obtained and specially preserved, i.e. not in existing human archives from major mass CRC screening and prevention studies. Our groundbreaking discoveries on genes that regulate PGE2 overcomes this challenge. Specifically, we have discovered that 15-PGDH, an enzyme that degrades PGE2, is: i) a major in vivo negative regulator of colon PGE2; ii) an effective surrogate biomarker of pan-colonic PGE2 biological activity; iii) a tumor suppressor gene, that, as we published in Science, iv) potently regulates proliferation of colon crypt stem cells, and whose deficiency we v) have linked to increased CRC risk. Moreover, in groundbreaking studies, published in Science Translational Medicine, we vi) have shown inter-individual differences of colon 15-PGDH range up to 12-fold, and vii) that aspirin works to lower CRC risk only in those who have high (above average) colon 15-PGDH (i.e. reducing colon PGE2 sufficiently to lower CRC risk requires a COX inhibitor plus robust colon 15-PGDH activity). Our key hypothesis is that by using the full PGE2 metabolic pathway we can develop an even more powerful predictor, based on comprehensively integrating expression of 6 genes that together regulate colon PGE2 degradation (15-PGDH and PGT) plus colon PGE2 synthesis (COX-1, COX-2, mPGES-1, and ABCC4). We hypothesize that this 6-gene signature of inferred PGE2 will (as compared to 15-PGDH alone) provide: i) an even stronger signature for identifying individuals with high colorectal cancer risk, who will benefit from more intensive screening; ii) an even stronger signature for identifying individuals who will benefit from using aspirin to prevent colorectal cancer; and iii) a target for identifying lifestyle interventions able to shift the colon PGE2 metabolic pathway toward an aspirin sensitive profile, thereby rendering added individuals sensitive to aspirin chemoprevention.
Our specific aims (Fig. 1) are:
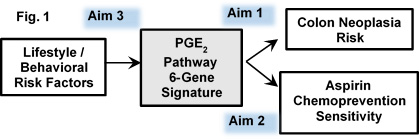
Aim 1. We will deliver a signature of colon neoplasia risk based on integrated assay of expression of a set of 6 key genes that regulate colon PGE2, and will show this 6-gene signature (inferred PGE2) is more predictive of risk than is colon 15-PGDH alone, interrogating this 6-gene panel in normal colon samples from 1922 individuals determined as having: normal colonoscopies, colon adenomas, or colon cancers.
Aim 2. We will test that the 6-gene expression signature of inferred PGE2 will identify individuals who are responsive to aspirin chemoprevention. We will further show this integrative 6-gene signature is more predictive of aspirin efficacy than is assay of colon 15-PGDH alone. We will test these hypotheses in colon mucosa samples from the Nurses' Health Study, the Health Professionals Follow-up Study, and the ACS Cancer Prevention Study-II.
Aim 3. We will test the hypothesis that specific lifestyle factors that both impinge on inflammatory tone and alter CRC risk act via regulating genes of the colon PGE2 pathway. Step 1 will test for association of obesity, physical activity, red meat consumption, and other CRC risk factors, to human colon expression of the 6 PGE2-pathway genes. Step 2 will use mouse models to functionally test lifestyle factors for ability to: i) regulate colon expression of the 6 PGE2 pathway genes; ii) shift the 6-gene expression signature of inferred PGE2 from an "aspirin resistant" to an "aspirin sensitive" profile; iii) induce the predicted parallel decrease of colon PGE2.
Project 2: Translational Significance of a Mutational Signature of African American Colon Cancers
Project Co-Leaders:
J. Willis, MD
Z. Wang, PhD
L. Li MD, PhD
African Americans (AAs) are more likely to be diagnosed with, and to die of, colorectal cancer than any other ethnic or racial group. Mortality rate for Caucasians has declined by 39% but increased by 28% for AAs since 1960, and the incidence in AAs remains 15 to 23% higher than Caucasians and other racial groups. The complex contributions of socio-economic disparities vs biological differences to the striking racial disparities are largely unknown. Our team has recently completed the first genome-wide analysis of the mutational landscape of colorectal cancers specifically arising in AAs. This landmark study identified 20 candidate driver genes never before seen in colorectal cancer and found that 15 of these mutations showed an over 3-fold increased mutation rate in AA colorectal cancers. These results identified that 41% of AA colorectal cancers have somatic mutations in at least one of the 15 genes, which is significantly different from colorectal cancers derived from Caucasians. Furthermore, 14% of AA colorectal cancers harbor mutations seen exclusively in AAs, most prominently including the ephrin type A receptor 6 gene [EPHA6]. Moreover, our recent follow up analysis showed that patients bearing the 15-gene mutational signature have worse clinical outcomes. These findings support our hypothesis that inherent biological differences may in part be responsible for the disparate burden of colorectal cancers in AAs. We further hypothesize that African ancestry, alone or in combination with environmental factors, is directly linked to the mutational signature we have identified in 41% of AA colorectal cancers. This project will fully investigate these concepts and define the functionality of EPHA6 mutations identified in AA colorectal cancers.
Aim 1: Validate the association of African ancestry with the 15-gene mutational signature in AA colorectal cancers and define the role of these mutations in AA colorectal cancer progression. Aim 1a: To validate and establish the generality of the 15-gene AA colorectal cancer mutational signature by re-sequencing a large independent set of AA colorectal cancers (along with a comparison set of European American colorectal cancers). Aim 1b: To determine whether African ancestry is genetically linked to these mutations by first determining if these somatic mutations are selectively targeted to develop on African inherited alleles and, secondly by testing if these mutations also develop in colorectal cancers from Nigeria, a population with a 100% African genome. Aim 1c: To investigate where in the process of cancer initiation and progression these mutations occur by comparing mutational signatures in adenomas vs adenocarcinomas. Aim 1d: To examine whether the mutational signature is linked to distinct clinicopathological features. In support of this inquiry, our preliminary data show that all 7 AA colorectal cancers harboring EPHA6 mutations are located in the right colon.
Aim 2: Assess survival and risk association with AA colorectal cancer mutations. Aim 2a: To determine the clinical significance of the mutational signature by assessing whether disease-free survival of AA colorectal cancers harboring mutations of the signature genes differ from those without mutations. Aim 2b: To assess whether environmental and lifestyle risk factors are differentially associated with subtypes of AA colorectal cancer defined by the 15-gene mutations. In particular, we will test the hypothesis that obesity, red meat intake, smoking, alcohol, and physical activity are differentially associated with subtypes of AA colorectal cancer. These factors are well established in the etiology of colorectal cancer and have been implicated in the observed racial disparities. We will further explore whether other lifestyle factors are also differentially associated with mutation subtypes of colorectal cancers in AAs. We will leverage the Southern Community Cohort Study [SCCS] and the North Carolina Colon Cancer Study [NCCCS], where epidemiological data and colonic tissues have been collected and are readily available.
Aim 3: Determine functional consequences of AA colorectal cancer-derived EPHA6 mutations. EPHA6 is the most frequently mutated gene in the 15 gene signature; and is mutated exclusively in AA colorectal cancers. We identified splice-site and frame-shrift mutations of EPHA6 that result in premature truncation of the protein. These data suggest that EPHA6 normally functions as a tumor suppressor. We will test this hypothesis by determining whether: EPHA6 suppresses cell growth in culture, anchorage independent growth, migration, invasion, and xenograft tumor formation using both subcutaneous and orthotopic colon cancer models (Aim 3a). To this end, we will re-express WT EPHA6 in two colon cancer cell lines that harbor EPHA6 loss-of-function mutations; EPHA6 mutations impact intestinal cell fate and promote tumor formation in innovative human intestinal organoid models by knocking in EPHA6 mutations using cutting-edge CRISPR/Cas9 technology (Aim 3b); and EPHA6 knockout mice increase number or size of intestinal tumors in Apc+/min genetic background (Aim 3c).
Significance and innovation: The success of this study will: i) establish a molecularly defined subtype of colorectal cancer that uniquely arises in AA individuals; ii) will elucidate the biological basis for the development of the mutations that define these cancers; and iii) will determine the impact this subtype of colorectal cancer has on disease outcome disparities in AAs. The study will inform new approaches to diagnosis, prevention and potentially treatment of AA colorectal cancers. This innovative study will establish a new paradigm to understand colorectal cancer disparities facing the AA community. Moreover, this study will utilize innovative human intestinal organoid models. This project meets the designation of a SPORE Population Sciences Project.
Project 3: Molecular Markers for Early Detection of Esophageal Carcinogenesis
Project Co-Leaders:
A. Chak, MD
Sanford D. Markowitz, MD, PhD
K. Guda, DVM, PhD
Prospectively collected Surveillance Epidemiology and End Results (SEER) data indicate that the incidence of esophageal adenocarcinoma (EAC) has increased more than 5-fold in the past three decades. Over 10,000 cases are now diagnosed annually. The prognosis for patients with EAC is poor with less than 20% of patients surviving beyond 5 years.
Barrett's esophagus (BE), a pre-malignant metaplastic condition diagnosed only when patients undergo EGD, is the only known precursor of EAC. However, due to the high cost of EGD and the lack of a randomized controlled trial supporting its efficacy, endoscopy to screen for BE is not routinely recommended. Thus, the previous presence of BE is unknown in over 95% of EAC cases. Most BE remains undetected. Clearly, to impact on the dismal prognosis of EAC the first step must be to develop alternative methods for identifying the at risk population who have BE. This will require new methods that are less expensive than EGD and can be widely accepted and adopted in a population at risk. Our discovery of methylated vimentin (mVIM) plus other robust methylated DNA biomarkers leads us to now propose a non-endoscopic method for early detection of BE.
Detection of BE alone will not fully solve the challenge. Once BE has been diagnosed, subsequent surveillance with periodic EGD is the current strategy for the early detection of dysplasia/cancer. However, most patients with BE do not progress to EAC and surveillance may or may not result in improved survival. Thus, not only is it imperative to first develop effective screening methods to detect BE, it is equally imperative to then develop novel surveillance strategies to detect progression of BE to high grade dysplasia (HGD) and frank EAC. This will require developing a panel of novel biomarkers that can detect the early progression of BE to HGD and EAC.
More recently, using reduced representation bisulfite sequencing (RRBS) and RNA sequencing (RNA-seq), we identified novel candidate molecular markers (methylated DNA and RNAs) for both BE screening and surveillance. Accordingly, in this proposal, we will evaluate a two-step biomarker based strategy. Step one is envisioned as biomarker based non-endoscopic BE detection and step two is biomarker guided selective BE surveillance. Furthermore, we have also explored whether these BE and EAC associated biomarkers have a functional role in BE carcinogenesis. None of the methylated DNA biomarkers we discovered were associated with epigenetic silencing of gene expression. Our proposal meets the designation of a SPORE Early Detection Project whose Specific Aims are:
Specific Aim 1. To develop a non-endoscopic methylation biomarker based method for early detection of BE. We propose a pilot clinical trial to determine whether we can detect BE in patients using mVIM plus other methylated markers and a novel non-endoscopic balloon-based esophageal sampling device we have designed.
Specific Aim 2. To validate candidate biomarker panels associated with progression from BE to EAC. We will determine the best set of progression markers. To do this we will validate a molecular marker panel comprised of: novel progression associated methylated CpG patches plus p53 mutations plus marker RNAs in a large independent cohort of normal, BE, and EAC tissues. These biomarker panels will be sequentially tested on biopsies, on endoscopic esophageal brushings, and finally in biospecimens obtained with the balloon device.
Specific Aim 3. To elucidate the biological function of EAC associated RNAs in malignant transformation. We have identified two RNAs that are markedly overexpressed in EACs compared to BE. We will employ cutting-edge gene targeting and integrative molecular profiling approaches to dissect the functional roles of these candidate onco-RNAs. Specifically, we will determine the phenotypic effects of these RNA markers on tumor growth and malignant phenotype. We further will delineate the critical downstream pathway networks modulated by each of the candidate RNAs.
These studies, once successful, will not only lead to the development of evidence-based molecular early detection and surveillance strategies, but will also provide novel targets for effective prevention and treatment.
Project 4: Targeting Glutamine Metabolism in Colorectal Cancer Harboring PIK3CA Mutations
Project Co-Leaders:
Z. Wang, PhD
D. Bajor, MD
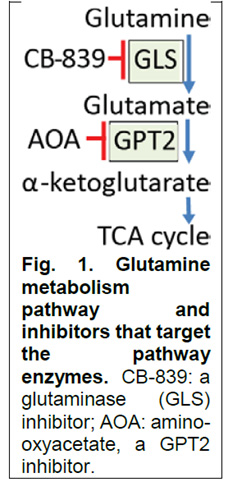
This proposal is based on our novel discovery that colorectal cancers (CRCs) with PIK3CA mutations are addicted to glutamine (Nature Communications, 7:11971, 2016), with growth of PIK3CA mutant CRC xenografts suppressed by inhibitors of glutaminase (CB- 839) or inhibitors of glutamine pyruvate transaminase 2/GPT2 (AOA) (Fig.1). In contrast, the two inhibitors have no effect on CRC xenografts with WT PIK3CA. Moreover, we find that the combination of CB-839 plus 5-fluorouracil (5-FU) is able, in 3 different models, to shrink PIK3CA mutant CRCs that grow through 5-FU alone. Remarkably, a third of these tumors were completely cured. These exciting results enabled a collaboration between Calithera Bioscience, the company that developed CB-839, and Dr. Neal Meropol (clinical PI of this project) to perform phase I and II clinical trials of the drug combination. The basic science PI, Dr. Zhenghe Wang, co-discovered that PIK3CA is highly mutated in many human cancers, including ~ 20% of CRCs (Science, 304:554,2004). We now find that PIK3CA mutations render CRC addicted to glutamine through up-regulation of GPT2, resulting an increased production of a-KG to replenish the tricarboxylic acid (TCA) cycle (Fig. 1). The overarching goal of this project is to exploit this dependence to develop a novel, mechanism-based treatment for patients with PIK3CA mutant CRCs. This will be accomplished by: i) defining the mechanisms by which inhibition of glutamine metabolism sensitizes PIK3CA mutant CRCs to 5-FU; ii) exploring the pharmacodynamics and antitumor activity of CB-839 in CRC patients; and iii) developing GPT2-specific inhibitors, a downstream target in the glutamate pathway.
Aim 1. Identify the mechanisms by which CB-839 augments the activity of 5-FU in PIK3CA mutant CRCs. Firstly, to solidify our preliminary results that CB-839 preferentially inhibits xenograft tumor growth of PIK3CA mutant CRCs, we will determine the tumor inhibitory effect of CB-839 against a panel of CRC patient-derived xenografts (PDXs) using orthotopic colon cancer models, as well as in an oncogenic PIK3CA mutant knockin mouse model. Secondly, we will define the mechanism by which CB-839 augments the activity of 5-FU. Our preliminary data indicate that CB-839 treatment increases expression of uridine phosphorylase 1 (UPP1) in PIK3CA mutant CRC cells. Knockout of UPP1 by CRSPR/Cas9 abrogates the ability of the combination of CB- 839 with 5-FU to shrink xenograft tumors. Given that UPP1 metabolizes 5-FU and facilitates its incorporation into DNA and RNA, we will determine: (1) if CB-839 treatment increases 5-FU incorporation into DNA and RNA through UPP1; and (2) how CB-839 treatment induces UPP1 gene expression. Thirdly, we will determine if the combination of CB-839 with 5-FU in xenograft tumors: (i) reduces proliferation, (ii) increases apoptosis, (iii) increases autophagy and/or (iv) reduces angiogenesis.
Aim 2. Assess the pharmacodynamic effects and clinical activity of CB-839 in CRC patients with PIK3CA mutant tumors. In collaboration with Calithera Bioscience, we have just opened a phase 1 trial to determine the recommended phase 2 doses of CB-839 plus capecitabine, an oral prodrug of 5-FU. Continuing this partnership, herein we propose to conduct a phase II clinical trial to determine the antitumor activity and pharmacodynamic effects of CB-839 plus capecitabine in patients with metastatic CRCs that harbor PIK3CA mutations and are resistant to prior fluoropyrimidine therapy. With serial tumor biopsies, we will test the hypothesis that CB-839 results in reduced glutaminase activity in tumors, induction of UPP1 gene expression in tumors and alteration of other biomarkers that will be informed by the studies performed in Aim 1. We also hypothesize that CB-839 in combination with capecitabine will overcome prior fluoropyrimidine resistance.
Aim 3. Develop more potent and specific GPT2 inhibitors. We found that shRNA knockdown of GPT2 abrogates the capacity of PIK3CA mutant CRC cells to form tumors, suggesting that GPT2 is also a critical target in these glutamine addicted CRCs. Consistent with this hypothesis, we find that aminooxyacetate (AOA), a pan- transaminase inhibitor, selectively inhibits the growth of xenografts of CRCs with PIK3CA mutations. Moreover, sensitivity to AOA is lost in GPT2 knockdown cells, indicating GPT2 is the target of AOA. These results led us to hypothesize that GPT2-specific inhibitors will be more potent and less toxic than AOA, and thus would have a more favorable therapeutic index than either AOA or CB-839. To test this hypothesis, we will develop potent and selective inhibitors of GPT2 by using structure-guided design to optimize our initial hit, AOA. Optimized inhibitors will provide tool compounds to validate GPT2 as a viable cancer target. Potency and selectivity will be determined by assaying IC50 against GPT2 and closely related enzymes. Potent and specific GPT2 inhibitors will be tested for tumor inhibitory effect on PIK3CA mutant CRCs using tissue culture and xenograft tumor models.
Significance and innovation: PIK3CA is the most frequently mutated oncogene in human cancers. However, effective approaches to treat patients with PIK3CA mutant tumors have not yet been developed. We propose to investigate an innovative concept of targeting glutamine addiction in CRCs with PIK3CA mutations. The success of this study will: 1) develop a novel precision therapy; 2) elucidate the molecular mechanisms by which CB-839 augments the therapeutic effect of fluoropyrimidines; and 3) develop novel compounds that potently inhibit GPT2. Moreover, this study utilizes cutting-edge CRSPR/Cas9 genome editing technology and innovative orthotopic colon cancer PDX models.
Administrative Core
Core Directors:
Sanford D. Markowitz, MD, PhD
N. Berger, MD
The Administrative Core is responsible for overall leadership and management of the Case GI SPORE. It will provide for overall direction, coordination, and administration of the GI SPORE. It will be responsible for facilitating, coordinating, stimulating, monitoring and maintaining scientific and financial oversight for all SPORE activities. It will provide the central forum for interaction among all SPORE components and all SPORE faculty. It will provide the central mechanism for monitoring progress and evaluating performance of all SPORE components. It will provide the coordinating mechanism between the SPORE faculty and the SPORE Internal and External Advisory Boards. And it will provide the mechanism for one of the key functions of the SPORE, the decision on when to re-allocate SPORE funds, in recognition of success of or roadblocks encountered by individual SPORE projects, or in order to recognize and take advantage of new scientific opportunities. The Administrative Core will maintain liaison between the GI SPORE, the Case Comprehensive Cancer Center, and the School of Medicine leadership, as well as with external organizations including NCI, other GI SPOREs, and the SPORE community in general.
The Administrative Core will include: The SPORE Principal Investigator, the SPORE Co-Principal Investigator, and the SPORE Administrative Coordinator. The Administrative Core will oversee the functioning of: the SPORE Executive Committee, the SPORE Internal Advisory Board, the SPORE External Advisory Board, and the SPORE Patient Advocates Advisory Board.
The specific aims of the core will be to:
Aim 1. Provide for interaction and scientific exchange among the SPORE faculty and between the SPORE faculty and the Case Comprehensive Cancer Center and the Case School of Medicine communities.
Aim 2. Provide a mechanism for evaluation and monitoring of the progress of all SPORE components.
Aim 3. Provide administrative support to enable the workings of all SPORE Research Projects, SPORE shared resources, the SPORE Career Development Program, and the SPORE Developmental Research Program.
Aim 4. Provide a mechanism for financial administration of the SPORE.
Aim 5. Provide a mechanism to assure interaction and integration of the SPORE with the Case Comprehensive Cancer Center and the Case School of Medicine.
Aim 6. Provide central oversight for data management activities of the SPORE.
Aim 7. Assure SPORE compliance with all federal and institutional regulations and reporting requirements.
Specific means by which the Administrate Core will achieve these aims will include:
- Convene regular monthly meetings of SPORE Executive Committee;
- Provide for twice yearly oral progress presentations to the Executive Committee from all SPORE components;
- Provide for annual oral progress presentations from all SPORE components at the annual SPORE retreat;
- Provide for annual written progress reports for all SPORE components;
- Convene an annual meeting of the SPORE Internal Advisory Board for formal review of all SPORE component reports and for formal evaluation and scoring of the success of each component;
- Convene an annual meeting of the SPORE External Advisory Board for formal review of all SPORE component reports and for formal evaluation and scoring of the success of each component;
- Convene an annual meeting of the Patient Advocates Board for input and review of SPORE activities;
- Coordinate solicitation and evaluation of pilot research studies for the Developmental Research Program;
- Coordinate solicitation for and evaluation of Career Enhancement Program awardees;
- Oversee conduct of all SPORE activities;
- Oversee administration and coordination of SPORE Core Facility utilization and accounting;
- Oversee quality control and quality improvement processes for all SPORE Core Facilities;
- Oversee management and accounting of grant funds and assure monthly reconciliation of all accounts;
- Assure compliance and maintain documents for all institutional regulatory requirements for human experimentation, animal utilization, toxic material and safety training and utilization;
- Oversee compliance and coordination with all reporting regulations;
- Oversee compliance with all requirements for research progress reports;
- Oversee compliance with NIH public access (PMCID) policies;
- Assure participation of the Case GI SPORE faculty in NCI sponsored meetings and workshops;
- Oversee and prioritize use of SPORE discretionary funds;
- Manage all intellectual property affairs arising from SPORE-sponsored research;
- Coordinate SPORE seminars and retreats;
- Provide central oversight of all data management activities of the SPORE Biostatistics and Informatics Core;
- Represent the SPORE in interactions with other Centers, Departments, the School of Medicine, affiliated hospitals and community organizations;
- Assure continuity of SPORE leadership.
Biospecimen Core
Core Director:
J. Willis, MD
The Biospecimen Core plays a central role in all GI SPORE projects and is critical to the success of the SPORE. This Core leverages the existing resources of the Case Comprehensive Cancer Center [Case CCC] Tissue Management Resource (which include Tissue Procurement, Histology, and Immunohistochemistry), but adds substantial SPORE specific capabilities, such as tracking and annotation of long term clinical outcomes linked to GI tissues, accessioning of the University Hospitals Case Medical Center [UHCMC] formalin fixed paraffin embedded [FFPE] tissue archive; as well as linkage with UHCMC’s large routine and molecular testing laboratories, for translational research purposes, clinical trials support expertise of the Case CCC Translational Research & Pharmacology Core Facility, and faculty level support for the experimental pathology needs of SPORE translational research studies.
Based on many years of experience in tissue based translational research, Core personnel interact with SPORE investigators in all stages of research, beginning with the formulation of research questions, through experimental design, developing unique high quality specimen datasets, data generation and interpretation, interaction with other Case Cores and entities to ensure sample transfer and characterization, sample tracking through project completion, culminating with writing scientific manuscripts and grants, as well as dissemination of results. As is illustrated in the discussion and figures of the Resource Service Plan, protocol modification and technology development at the Biospecimen Core are dictated by the needs of the GI SPORE Projects and by close and frequent interaction of Biospecimen Core personnel with SPORE Project leaders.
SPECIFIC AIMS OF THE BIOSPECIMEN CORE:
Aim 1. To supply quality controlled human tissue biospecimens, including both frozen and formalin fixed paraffin embedded (FFPE) tissues, annotated with long-term clinical follow-up and clinical outcomes, which will enhance GI cancer translational research studies.
Aim 2. To provide access to a full range of expert tissue based technologies (including microdissection, laser capture microdissection, tissue microarray, and development of custom immunohistochemistry and in situ hybridization assays), which enable the GI SPORE studies proposed in this application.
Aim 3. To provide support and expertise to Case GI SPORE investigators in the design and implementation of tissue based research projects — including new techniques and technologies as required.
Aim 4. To acquire specific quality controlled specialized tissue samples from outside institutions and make them efficiently available to Case GI SPORE investigators.
Aim 5. To offer comprehensive human GI pathology consultation and experimental pathology consultation, which will enhance the research studies carried out by GI SPORE investigators.
OVERVIEW OF THE BIOSPECIMEN CORE CAPABILITIES
Sample Acquisition
- Comprehensive acquisition and banking of fresh tissues — including neoplasia and controls and other specialized tissues — such as esophageal mucosal samples.
- Collection and processing of tissues and blood samples specifically for SPORE research purposes.
- Collection of biospecimens from participants in SPORE clinical trials.
- Collection of thousands of clinically derived FFPE samples from UHCMC Department of Pathology.
- Interaction with other SPOREs [and non-SPORE centers] to acquire tissues and expertise.
Sample Annotation
- Annotation of clinicopathological information linked to tissues including specialized molecular test results and patient follow-up into a centralized database via IRB approved standard operating procedures insuring patient confidentiality.
Morphological Review and Dissection
- Quality control of all tissue samples used for GI SPORE research.
- Morphological review and interpretation of human and mouse GI tissues used in SPORE research.
- Tissue microdissection including laser capture microdissection.
Down-Stream Tissue Processing
- Tissue microarray construction and analyses.
- Processing of tissues for research purposes including FFPE and frozen sections.
- Immunohistochemistry and immunoflourescence including multicolor procedures and analyses.
- In situ hybridization.
- RNA/DNA extraction from FFPE tissue and blood for investigators who lack these capabilities.
- Blood fractionations.
Pathology Consultation
- Provide a broad range of pathology expertise including insights to current issues in clinical medicine relating to proposed SPORE research projects.
Biostatistics and Informatics Core
Core Director:
J. Barnholtz-Sloan, PhD
The Case GI SPORE Biostatistics and Informatics Core (BIC) will interact with all SPORE investigators to develop and apply appropriate analysis methods, study designs and secure databases to most efficiently achieve project goals. We will work closely with investigators to ensure that research questions lead to experiments with quantifiable goals and testable hypotheses. In addition, the Case GI SPORE BIC will provide infrastructure and oversight for secure management of all study data, working closely with the Biospecimen Core to facilitate accuracy and completeness of datasets for all projects and investigators.
Specific Aims are:
Aim 1. To provide support to Case GI SPORE investigators in the design, conduct, analysis and reporting of results from Case GI SPORE studies
Aim 2. To provide secure and accessible database systems to facilitate reliable and reproducible impactful results for all Case GI SPORE investigators
The Biostatistics and Informatics Core (BIC) will serve as a centralized resource for all investigators involved in the Case GI SPORE, including the leaders of the 4 translational research projects, Developmental Research Program (DRP) investigators, Career Enhancement Program (CEP) investigators and leaders of the Biospecimen Core. The Core will assist investigators with all facets of statistical design, analysis and interpretation. Established as well as novel tailored analytic methods are available to promote scientific discovery for GI cancers within the Core. Core personnel will interact with SPORE investigators in all stages of research, from the formulation of the research questions, through experimental design, data management, data analysis (including outcomes analysis), interpretation, writing scientific manuscripts and grants, to dissemination of results. The Core will also facilitate access to new or enhancement of existing secure and accessible database systems in order to assist with statistical analysis, discovery and collaboration. The Core will assist all investigators with development, management and reliability of their project data and will also integrate information across databases, such as incorporating clinical and experimental data with biospecimen data as provided by the Biospecimen Core. The BIC will provide a natural foundation for cross-pollination among the projects and Cores, the Core personnel collaborating with all project investigators and Core directors.
The BIC leverages faculty and resources in the Case Comprehensive Cancer Center, Case Western Reserve University (CWRU) School of Medicine and the Institute for Computational Biology (ICB) at CWRU. Dr. Jill Barnholtz-Sloan, Co-Director of the SPORE Biostatistics Core, also serves as Associate Director of Bioinformatics in the Case Comprehensive Cancer Center (Case CCC) and as a co-Director of the Case CCC Biostatistics and Bioinformatics Core Facility (BBCF). Dr. Barnholtz-Sloan is thus perfectly positioned to assure that decisions and investments made by the SPORE Biostatistics Core will be complementary to and will leverage resources available in the Case CCC Biostatistics and Bioinformatics Core. Moreover, the Biostatistics and Bioinformatics Environment has been at Case has been markedly enhanced with the recent recruitment of Dr. Jonathan Haines as Chairman of the Case Department of Quantitative Health Sciences and as Director of the Institute of Computational Biology (ICB), a major initiative between Case and its allied hospitals (University Hospitals Case Medical Center and the Cleveland Clinic) to facilitate translational research by providing a unified platform for data security and data availability for clinical research. The ICB will be developing integrated programs of support for the Case CCC. Within the ICB Dr. Haines holds the position of ICB Director and Dr. Barnholtz-Sloan holds the position of Associate Director for Translational Informatics. As Co-Directors of the SPORE Biostatistics and Informatics Core, Drs. Haines and Barnholtz-Sloan are thus perfectly positioned to assure that decisions and investments made by the SPORE Biostatistics Core will be complementary to and will leverage the resources of the newly formed ICB. Drs. Barnholtz-Sloan and Haines also have the quantitative skills and leadership abilities to ensure success of the Core and the Case GI SPORE investigators, projects, Cores, CEP and DRP awardees.
Developmental Research Program
Program Director:
N. Berger, MD
The Case GI SPORE values the critical contribution of innovative and diverse approaches to the future of GI translational research and is strongly committed to provide 1) financial support, 2) core facility services from the SPORE, the Comprehensive Cancer Center and participating institutions and 3) intellectual oversight and advice through the GI SPORE Developmental Research Program (DRP). Accordingly, emphasis will be placed on supporting new investigators as well as established investigators with new approaches to GI cancers. The Case GI SPORE considers the developmental project mechanism to be an important opportunity for high risk/high impact research and will provide appropriate prioritization for such proposals. The overall goal of the DRP is to stimulate new ideas and new approaches to reduce the incidence and mortality rate of GI cancers. Plans are included to expand the focus of these pilot research programs to GI Malignancies beyond colorectal cancer, the traditional focus of research excellence at Case Western Reserve University (CWRU). Demonstrating commitment to the DRP, at both the institution and SPORE level, the following annual commitments have been made to support Pilot Project Research: School of Medicine, $50,000, Case Comprehensive Cancer Center $50,000 per year to be applied flexibly to either DRP or Career Enhancement Program, Cleveland Clinic Tausig Cancer Center $50,000, that are all in addition to $50,000 from SPORE Grant funds, thus providing support for at least 3 and potentially 4 pilot projects, at $50,000 each, per year, each year of SPORE funding. We anticipate the opportunities created by these pilot projects will continue to attract widespread interest from throughout the Case Campus and affiliated hospitals, contribute to our accelerated momentum in GI cancer research, expand our portfolio in GI cancer research beyond colorectal cancer and provide, as has been demonstrated over the past 5 years, investigators with support to generate sufficient preliminary data to apply for individual NIH R01 grants or to be considered for future support as full SPORE Research Programs. In addition to this program engaging individual investigators from affiliated institution to participate in research programs, the DRP has and we expect will continue to encourage and enhance cooperation in clinical-translational research activities among our constituent institutions.
GOALS:
The Case GI SPORE will award at least 3 and potentially 4 Developmental Research Projects annually, at up to $50,000 each, to stimulate innovative development in Gastrointestinal Translational Cancer Research.
SPECIFIC AIMS OF THE SPORE DEVELOPMENTAL RESEARCH PROGRAM:
Aim 1. To provide support for innovative, translational developmental research projects across the spectrum of GI malignancies from etiology through prevention, screening, diagnostics, therapy and survivorship.
Aim 2. To encourage interest and engagement in translational GI malignancy research by investigators from diverse disciplines employing varied approaches including, biologic, molecular biologic, epidemiologic, genetic, omic, informatic, biomedical engineering, psychosocial, comparative effectiveness, basic, clinical and translational methodologies and technologies.
Aim 3. To stimulate novel ideas and to encourage application of innovative approaches from other disciplines and organ systems to focus on translational GI cancer research.
Aim 4. To demonstrate feasibility of translational research concepts and obtain preliminary data to support development of fully independent and/or full SPORE Research Projects.
Aim 5. To establish new collaborations with other SPORE and/or institutions in translational GI research.
Career Enhancement Program
Program Director:
N. Berger, MD
Building on a strong history of training and development of successful GI Cancer Researchers at Case Western Reserve University (CWRU), the overall objective of the Case GI SPORE Career Enhancement Program (CEP) is to develop the next generation of GI Cancer Translational Research Scholars who are, 1) trained in the broad fundamentals of GI cancer pathobiology; 2) ready to lead independent translational GI cancer research programs, 3) oriented and able to most effectively utilize innovative translational and transdisciplinary science approaches to attack and solve problems associated with GI malignancies, 4) committed to improving all aspects of patient health relevant to GI malignancies across the spectrum from etiology through prevention, screening, diagnosis, therapy and survivorship, and 5) prepared with sufficient understanding to effectively utilize academic, pharmaceutical, community and national resources to most effectively support translational research program development and promote academic advancement.
Specific Aims: The specific aims of the CEP are as follows:
Aim 1. To recruit, support and develop talented investigators, as SPORE Career Enhancement Program Scholars (CEP-S) to conduct translational research focused on GI cancers.
Aim 2. To provide outstanding translational research mentorship, training and opportunities for investigators to develop sufficient knowledge, skills and expertise to successfully pursue independent translationally oriented scholarly research careers focused on GI malignancies.
Aim 3. To provide broad career mentoring, and support development of research and organizational skills to ensure the CEP -S are optimally prepared to engage and benefit from the interaction with the institutional, academic, pharmaceutical, research, funding, regulatory and patient based communities.
Aim 4. To expand and enhance the cadre of investigators at CWRU focused on innovative GI Cancer Research and to develop new investigators with all the competencies required to successfully initiate and conduct independent R01 or SPORE type projects focused on GI cancer research.







